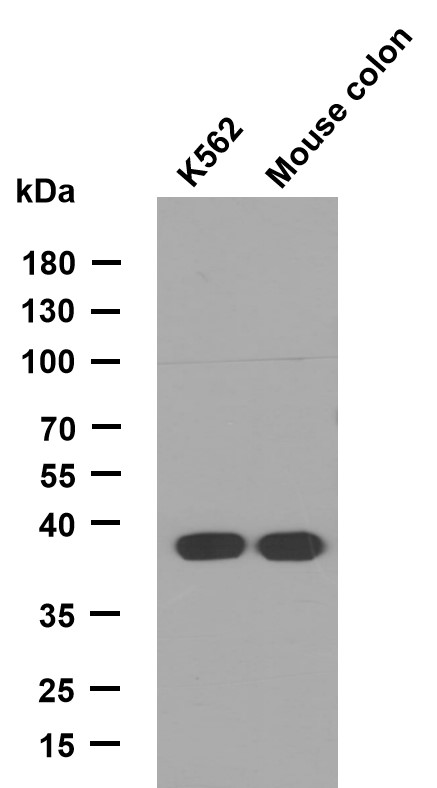
Histone H2A.X (Phospho Ser139) Polyclonal Antibody
- Catalog No.:YP1653
- Applications:WB
- Reactivity:Human;Mouse;Rat
- Target:
- Histone H2A.X
- Fields:
- >>Necroptosis;>>Neutrophil extracellular trap formation;>>Alcoholism;>>Systemic lupus erythematosus
- Gene Name:
- H2AFX
- Protein Name:
- Histone H2A.x,γH2AX
- Human Gene Id:
- 3014
- Human Swiss Prot No:
- P16104
- Mouse Gene Id:
- 15270
- Mouse Swiss Prot No:
- P27661
- Immunogen:
- Synthetic Peptide of Histone H2A.X (Phospho Ser139)
- Specificity:
- The antibody detects endogenous Histone H2A.X (PhosphoSer139) protein.
- Formulation:
- PBS, pH 7.4, containing 0.5%BSA, 0.02% sodium azide as Preservative and 50% Glycerol.
- Source:
- Polyclonal, Rabbit,IgG
- Dilution:
- WB 1:1000-2000
- Purification:
- The antibody was affinity-purified from rabbit antiserum by affinity-chromatography using specific immunogen.
- Storage Stability:
- -15°C to -25°C/1 year(Do not lower than -25°C)
- Other Name:
- H2AFX;H2AX;Histone H2A.x;H2a/x
- Observed Band(KD):
- 15kD
- Background:
- Histones are basic nuclear proteins that are responsible for the nucleosome structure of the chromosomal fiber in eukaryotes. Two molecules of each of the four core histones (H2A, H2B, H3, and H4) form an octamer, around which approximately 146 bp of DNA is wrapped in repeating units, called nucleosomes. The linker histone, H1, interacts with linker DNA between nucleosomes and functions in the compaction of chromatin into higher order structures. This gene encodes a replication-independent histone that is a member of the histone H2A family, and generates two transcripts through the use of the conserved stem-loop termination motif, and the polyA addition motif. [provided by RefSeq, Oct 2015],
- Function:
- developmental stage:Synthesized in G1 as well as in S-phase.,domain:The [ST]-Q motif constitutes a recognition sequence for kinases from the PI3/PI4-kinase family.,function:Variant histone H2A which replaces conventional H2A in a subset of nucleosomes. Nucleosomes wrap and compact DNA into chromatin, limiting DNA accessibility to the cellular machineries which require DNA as a template. Histones thereby play a central role in transcription regulation, DNA repair, DNA replication and chromosomal stability. DNA accessibility is regulated via a complex set of post-translational modifications of histones, also called histone code, and nucleosome remodeling. Required for checkpoint-mediated arrest of cell cycle progression in response to low doses of ionizing radiation and for efficient repair of DNA double strand breaks (DSBs) specifically when modified by C-terminal phosphorylation.,PTM:Mon
- Subcellular Location:
- Nucleus . Chromosome .
- Expression:
- Lung,Placenta,
- June 19-2018
- WESTERN IMMUNOBLOTTING PROTOCOL
- June 19-2018
- IMMUNOHISTOCHEMISTRY-PARAFFIN PROTOCOL
- June 19-2018
- IMMUNOFLUORESCENCE PROTOCOL
- September 08-2020
- FLOW-CYTOMEYRT-PROTOCOL
- May 20-2022
- Cell-Based ELISA│解您多样本WB检测之困扰
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-ACETYL-PROTEIN
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-PHOSPHO-PROTEIN
- July 13-2018
- Antibody-FAQs