- Home
- About
- Promotions
-
Products
-
Elisa Kits
- |
-
Primary antibodies
- |
-
Secondary antibodies
- |
-
Proteins
- |
-
IHC reagents
- |
-
WB reagents
- PonceauS Staining Solution
- PBST Washing Buffer, 10X
- 1.5M Tris-HCl Buffer, pH8.8
- 1M Tris-HCl Buffer, pH6.8
- 10% SDS Solution
- Prestained Protein Marker
- TBST Washing Buffer, 10X
- SDS PAGE Loading Buffer, 5X
- Stripping Buffered Solution
- Tris Buffer, pH7.4, 10X
- Total Protein Extraction Kit
- Running Buffer, 10X
- Transfer Buffer, 10X
- 30% Acr-Bis(29:1) Solution
- Tris电泳液速溶颗粒
- PBS(1X, premixed powder)
- TBS(1X, premixed powder)
- 快速封闭液
- 转膜液速溶颗粒
- Chemical reagents
- News
- Distributor
- Resources
- Contact
- Home
- >
- Info
- >
- Nrf2 protein
- >
- Go Back
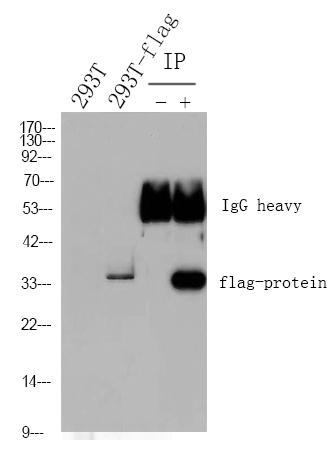
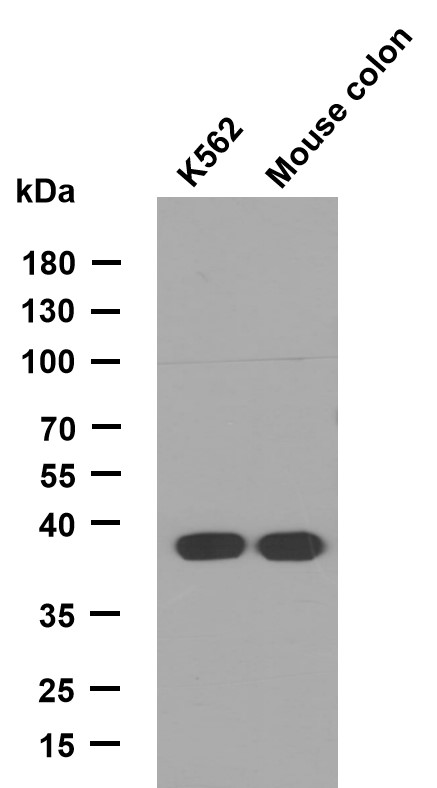
Nrf2 protein
- Catalog No.:YD0126
- Applications:WB;SDS-PAGE
- Reactivity:Human
- Protein Name:
- Nrf2 protein
- Sequence:
- Amino acid: 331-605, with his-MBP tag.
- Formulation:
- Liquid in PBS
- Concentration:
- SDS-PAGE >90%
- Storage Stability:
- -20°C/6 month,-80°C for long storage
- Other Name:
- Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (HEBP1) (Nuclear factor, erythroid derived 2, like 2)
- Background:
- domain:Acidic activation domain in the N-terminus, and DNA binding domain in the C-terminus.,function:Transcription activator that binds to antioxidant response (ARE) elements in the promoter regions of target genes. Important for the coordinated up-regulation of genes in response to oxidative stress. May be involved in the transcriptional activation of genes of the beta-globin cluster by mediating enhancer activity of hypersensitive site 2 of the beta-globin locus control region.,PTM:Phosphorylation of Ser-40 by PKC in response to oxidative stress dissociates NFE2L2 from its cytoplasmic inhibitor KEAP1, promoting its translocation into the nucleus.,similarity:Belongs to the bZIP family.,similarity:Belongs to the bZIP family. CNC subfamily.,similarity:Contains 1 bZIP domain.,subcellular location:Cytosolic under unstressed conditions, translocates into the nucleus upon induction by electrophilic agents.,subunit:Heterodimer. May bind DNA with an unknown protein. Interacts with KEAP1. Interacts via its leucine-zipper domain with the coiled-coil domain of PMF1.,tissue specificity:Widely expressed. Highest expression in adult muscle, kidney, lung, liver and in fetal muscle.,
- Function:
- transcription, transcription, DNA-dependent, regulation of transcription, DNA-dependent, transcription from RNA polymerase II promoter, ER-nuclear signaling pathway, response to unfolded protein, positive regulation of biosynthetic process, response to organic substance, positive regulation of macromolecule biosynthetic process,positive regulation of macromolecule metabolic process, positive regulation of gene expression, endoplasmic reticulum unfolded protein response, positive regulation of cellular biosynthetic process, RNA biosynthetic process, cellular response to stress, cellular response to unfolded protein, response to endoplasmic reticulum stress, regulation of transcription, positive regulation of transcription, DNA-dependent, positive regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process, positive regulation of transcription, regulation of embryon
- Subcellular Location:
- Cytoplasm, cytosol . Nucleus . Cytosolic under unstressed conditions: ubiquitinated and degraded by the BCR(KEAP1) E3 ubiquitin ligase complex (PubMed:15601839, PubMed:21196497). Translocates into the nucleus upon induction by electrophilic agents that inactivate the BCR(KEAP1) E3 ubiquitin ligase complex (PubMed:21196497). .
- Expression:
- Widely expressed. Highest expression in adult muscle, kidney, lung, liver and in fetal muscle.