- Home
- About
- Promotions
-
Products
-
Elisa Kits
- |
-
Primary antibodies
- |
-
Secondary antibodies
- |
-
Proteins
- |
-
IHC reagents
- |
-
WB reagents
- PonceauS Staining Solution
- PBST Washing Buffer, 10X
- 1.5M Tris-HCl Buffer, pH8.8
- 1M Tris-HCl Buffer, pH6.8
- 10% SDS Solution
- Prestained Protein Marker
- TBST Washing Buffer, 10X
- SDS PAGE Loading Buffer, 5X
- Stripping Buffered Solution
- Tris Buffer, pH7.4, 10X
- Total Protein Extraction Kit
- Running Buffer, 10X
- Transfer Buffer, 10X
- 30% Acr-Bis(29:1) Solution
- Tris电泳液速溶颗粒
- PBS(1X, premixed powder)
- TBS(1X, premixed powder)
- 快速封闭液
- 转膜液速溶颗粒
- Chemical reagents
- News
- Distributor
- Resources
- Contact
- Home
- >
- Info
- >
- HIF 1a protein
- >
- Go Back
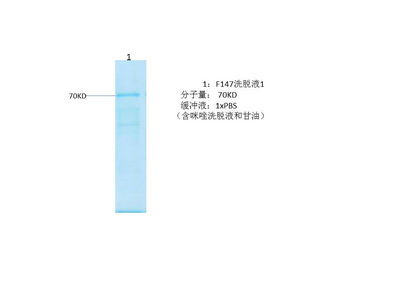
HIF 1a protein
- Catalog No.:YD0035
- Applications:WB;SDS-PAGE
- Reactivity:Human
- Protein Name:
- HIF 1a protein
- Sequence:
- Amino acid: 136-318, with his-MBP tag.
- Formulation:
- Liquid in PBS
- Concentration:
- SDS-PAGE >90%
- Storage Stability:
- -20°C/6 month,-80°C for long storage
- Other Name:
- Hypoxia-inducible factor 1-alpha (HIF-1-alpha) (HIF1-alpha) (ARNT-interacting protein) (Basic-helix-loop-helix-PAS protein MOP1) (Class E basic helix-loop-helix protein 78) (bHLHe78) (Member of PAS protein 1) (PAS domain-containing protein 8)
- Background:
- domain:Contains two independent C-terminal transactivation domains, NTAD and CTAD, which function synergistically. Their transcriptional activity is repressed by an intervening inhibitory domain (ID).,function:Functions as a master transcriptional regulator of the adaptive response to hypoxia. Under hypoxic conditions activates the transcription of over 40 genes, including, erythropoietin, glucose transporters, glycolytic enzymes, vascular endothelial growth factor, and other genes whose protein products increase oxygen delivery or facilitate metabolic adaptation to hypoxia. Plays an essential role in embryonic vascularization, tumor angiogenesis and pathophysiology of ischemic disease. Binds to core DNA sequence 5'-[AG]CGTG-3' within the hypoxia response element (HRE) of target gene promoters. Activation requires recruitment of transcriptional coactivators such as CREBPB and EP300. Activity is enhanced by interaction with both, NCOA1 or NCOA2. Interaction with redox regulatory protein APEX seems to activate CTAD and potentiates activation by NCOA1 and CREBBP.,induction:Under reduced oxygen tension. Induced also by various receptor-mediated factors such as growth factors, cytokines, and circulatory factors such as PDGF, EGF FGF-2 FGF-2 IGF-2, TGF-1 beta, HGF, TNF alpha, IL-1 beta, angiotensin-2 and thrombin. However, this induction is less intense than that stimulated by hypoxia.,online information:Hypoxia inducible factor entry,PTM:In normoxia, is hydroxylated on Asn-803 by HIF1AN, thus abrogating interaction with CREBBP and EP300 and preventing transcriptional activation. This hydroxylation is inhibited by the Cu/Zn-chelator, Clioquinol.,PTM:In normoxia, is hydroxylated on Pro-402 and Pro-564 in the oxygen-dependent degradation domain (ODD) by EGLN1/PHD1 and EGLN2/PHD2. EGLN3/PHD3 has also been shown to hydroxylate Pro-564. The hydroxylated prolines promote interaction with VHL, initiating rapid ubiquitination and subsequent proteasomal degradation. Under hypoxia, proline hydroxylation is impaired and ubiquitination is attenuated, resulting in stabilization.,PTM:Requires phosphorylation for DNA-binding.,PTM:S-nitrosylation of Cys-800 may be responsible for increased recruitment of p300 coactivator necessary for transcriptional activity of HIF-1 complex.,PTM:Sumoylated; by SUMO1 under hypoxia. Sumoylation is enhanced through interaction with RWDD3. Desumoylation by SENP1 leads to increased HIF1A stability and transriptional activity.,PTM:The iron and 2-oxoglutarate dependent 3-hydroxylation of asparagine is (S) stereospecific within HIF CTAD domains.,PTM:Ubiquitinated; in normoxia, following hydroxylation and interaction with VHL. Lys-532 appears to be the principal site of ubiquitination. Clioquinol, the Cu/Zn-chelator, inhibits ubiquitination through preventing hydroxylation at Asn-803.,similarity:Contains 1 basic helix-loop-helix (bHLH) domain.,similarity:Contains 1 PAC (PAS-associated C-terminal) domain.,similarity:Contains 2 PAS (PER-ARNT-SIM) domains.,subcellular location:Cytoplasmic in normoxia, nuclear translocation in response to hypoxia. Colocalizes with SUMO1 in the nucleus, under hypoxia.,subunit:Interacts with the HIF1A beta/ARNT subunit; heterodimerization is required for DNA binding. Interacts with COPS5; the interaction increases the transcriptional activity of HIF1A through increased stability (By similarity). Interacts with CREBBP and EP300 (via TAZ-type 1 domains). Interacts with NCOA1, NCOA2, APEX and HSP90. Interacts (hydroxylated within the ODD domain) with VHLL (via beta domain); the interaction, leads to polyubiquitination and subsequent HIF1A proteasomal degradation. During hypoxia, sumoylated HIF1A also binds VHL; the interaction promotes the ubiquitination of HIF1A. Interacts with SENP1; the interaction desumoylates HIF1A resulting in stabilization and activation of transcription. Interacts (Via the ODD domain) with ARD1A; the interaction appears not to acetylate HIF1A nor have any affect on protein stability, during hypoxia. Interacts with RWDD3; the interaction enhances HIF1A sumoylation. Interacts with TSGA10.,tissue specificity:Expressed in most tissues with highest levels in kidney and heart. Overexpressed in the majority of common human cancers and their metastases, due to the presence of intratumoral hypoxia and as a result of mutations in genes encoding oncoproteins and tumor suppressors.,
- Function:
- cell morphogenesis, cell morphogenesis involved in differentiation, angiogenesis, blood vessel development, response to hypoxia, ameboidal cell migration, in utero embryonic development, neural crest cell migration, regulation of cytokine production, positive regulation of cytokine production, epithelial to mesenchymal transition, placenta development, embryonic placenta development, regulation of endothelial cell proliferation, positive regulation of endothelial cell proliferation, vasculature development, healing during inflammatory response, connective tissue replacement during inflammatory response, lactate metabolic process, regulation of carbohydrate metabolic process,regulation of glycolysis, transcription, transcription, DNA-dependent, regulation of transcription, DNA-dependent,regulation of transcription from RNA polymerase II promoter, transcription from RNA polymerase II promo
- Subcellular Location:
- Cytoplasm . Nucleus . Nucleus speckle . Colocalizes with HIF3A in the nucleus and speckles (By similarity). Cytoplasmic in normoxia, nuclear translocation in response to hypoxia (PubMed:9822602). .
- Expression:
- Expressed in most tissues with highest levels in kidney and heart. Overexpressed in the majority of common human cancers and their metastases, due to the presence of intratumoral hypoxia and as a result of mutations in genes encoding oncoproteins and tumor suppressors. A higher level expression seen in pituitary tumors as compared to the pituitary gland.