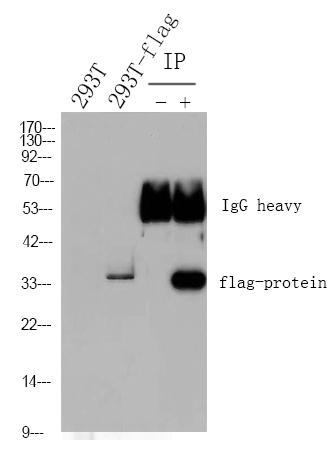
DNA pol β (Acetyl Lys72) rabbit pAb
- Catalog No.:YK0166
- Applications:WB;ELISA
- Reactivity:Human;Mouse;Rat
- Target:
- DNA pol β
- Fields:
- >>Base excision repair;>>Human T-cell leukemia virus 1 infection;>>Viral carcinogenesis
- Gene Name:
- POLB
- Protein Name:
- DNA pol β (Acetyl Lys72)
- Human Gene Id:
- 5423
- Human Swiss Prot No:
- P06746
- Mouse Gene Id:
- 18970
- Mouse Swiss Prot No:
- Q8K409
- Rat Gene Id:
- 29240
- Rat Swiss Prot No:
- P06766
- Immunogen:
- Synthesized peptide derived from human DNA pol β (Acetyl Lys72)
- Specificity:
- This antibody detects endogenous levels of Human,Mouse,Rat DNA pol β (Acetyl Lys72)
- Formulation:
- Liquid in PBS containing 50% glycerol, 0.5% BSA and 0.02% sodium azide.
- Source:
- Polyclonal, Rabbit,IgG
- Dilution:
- WB 1:1000-2000 ELISA 1:5000-20000
- Purification:
- The antibody was affinity-purified from rabbit serum by affinity-chromatography using specific immunogen.
- Concentration:
- 1 mg/ml
- Storage Stability:
- -15°C to -25°C/1 year(Do not lower than -25°C)
- Other Name:
- DNA polymerase beta (EC 2.7.7.7;EC 4.2.99.-)
- Observed Band(KD):
- 38kD
- Background:
- catalytic activity:Deoxynucleoside triphosphate + DNA(n) = diphosphate + DNA(n+1).,cofactor:Binds 2 magnesium ions per subunit.,domain:Residues 239-252 form a flexible loop which appears to affect the polymerase fidelity.,function:Repair polymerase. Conducts "gap-filling" DNA synthesis in a stepwise distributive fashion rather than in a processive fashion as for other DNA polymerases. Has a 5'-deoxyribose-5-phosphate lyase (dRP lyase) activity.,PTM:Methylation by PRMT6 stimulates the polymerase activity by enhancing DNA binding and processivity.,similarity:Belongs to the DNA polymerase type-X family.,subunit:Monomer.,
- Function:
- DNA metabolic process, DNA replication, DNA-dependent DNA replication, DNA repair, base-excision repair, pyrimidine dimer repair, anti-apoptosis, response to DNA damage stimulus, cell death, regulation of cell death, death, cellular response to stress, regulation of apoptosis, negative regulation of apoptosis, regulation of programmed cell death,negative regulation of programmed cell death, negative regulation of cell death,
- Subcellular Location:
- Nucleus. Cytoplasm. Cytoplasmic in normal conditions. Translocates to the nucleus following DNA damage.
- June 19-2018
- WESTERN IMMUNOBLOTTING PROTOCOL
- June 19-2018
- IMMUNOHISTOCHEMISTRY-PARAFFIN PROTOCOL
- June 19-2018
- IMMUNOFLUORESCENCE PROTOCOL
- September 08-2020
- FLOW-CYTOMEYRT-PROTOCOL
- May 20-2022
- Cell-Based ELISA│解您多样本WB检测之困扰
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-ACETYL-PROTEIN
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-PHOSPHO-PROTEIN
- July 13-2018
- Antibody-FAQs