SirT2 mouse mAb
- Catalog No.:YM1271
- Applications:WB;IF
- Reactivity:Human;Mouse;Rat
- Target:
- SirT2
- Fields:
- >>Nicotinate and nicotinamide metabolism;>>Metabolic pathways
- Gene Name:
- sirt2
- Human Gene Id:
- 22933
- Human Swiss Prot No:
- Q8IXJ6
- Mouse Swiss Prot No:
- Q8VDQ8
- Immunogen:
- Purified recombinant human SirT2 protein expressed in E.coli.
- Specificity:
- This antibody detects endogenous levels of SirT2 and does not cross-react with related proteins.
- Formulation:
- Liquid in PBS containing 50% glycerol, 0.5% BSA and 0.02% sodium azide.
- Source:
- Monoclonal, Mouse
- Dilution:
- wb 1:1000. IF 1:50-200
- Purification:
- The antibody was affinity-purified from mouse ascites by affinity-chromatography using epitope-specific immunogen.
- Concentration:
- 1 mg/ml
- Storage Stability:
- -15°C to -25°C/1 year(Do not lower than -25°C)
- Other Name:
- FLJ35621;FLJ37491;NAD dependent deacetylase sirtuin 2;NAD-dependent deacetylase sirtuin-2;NAD-dependent protein deacetylase sirtuin-2;Regulatory protein SIR2 homolog 2;Silencing information regulator 2 like;Silent information regulator 2;Silent mating type information regulation 2;Silent mating type information regulation 2 homolog;SIR 2;SIR2;SIR2 like;SIR2 like protein 2;Sir2 related protein type 2;SIR2, S. cerevisiae, homolog-loke 2;SIR2-like protein 2;SIR2L;SIR2L2;SIRT 2;SIRT2;SIRT2_HUMAN;Sirtuin (silent mating type information regulation 2 homolog) 2 (S.cerevisiae);Sirtuin 2;Sirtuin type 2;Sirtuin2.
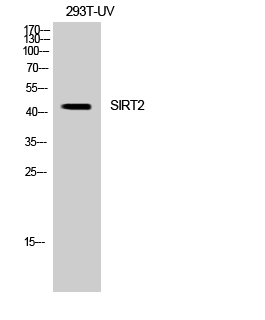
- Observed Band(KD):
- 43kD
- Background:
- This gene encodes a member of the sirtuin family of proteins, homologs to the yeast Sir2 protein. Members of the sirtuin family are characterized by a sirtuin core domain and grouped into four classes. The functions of human sirtuins have not yet been determined; however, yeast sirtuin proteins are known to regulate epigenetic gene silencing and suppress recombination of rDNA. Studies suggest that the human sirtuins may function as intracellular regulatory proteins with mono-ADP-ribosyltransferase activity. The protein encoded by this gene is included in class I of the sirtuin family. Several transcript variants are resulted from alternative splicing of this gene. [provided by RefSeq, Jul 2010],
- Function:
- catalytic activity:NAD(+) + an acetylprotein = nicotinamide + O-acetyl-ADP-ribose + a protein.,cofactor:Binds 1 zinc ion per subunit.,developmental stage:Peaks during mitosis. After mitosis, it is probably degraded by the 26S proteasome.,enzyme regulation:Inhibited by Sirtinol, A3 and M15 small molecules. Inhibited by nicotinamide.,function:NAD-dependent deacetylase, which deacetylates the 'Lys-40' of alpha-tubulin. Involved in the control of mitotic exit in the cell cycle, probably via its role in the regulation of cytoskeleton. Despite some ability to deacetylate histones in vitro, it is unlikely in vivo.,PTM:Phosphorylated at the G2/M transition of the cell cycle.,similarity:Belongs to the sirtuin family.,similarity:Contains 1 deacetylase sirtuin-type domain.,subcellular location:Colocalizes with microtubules.,subunit:Interacts with HDAC6, suggesting that these proteins belong to a la
- Subcellular Location:
- Nucleus . Cytoplasm, perinuclear region . Cytoplasm . Cytoplasm, cytoskeleton . Cytoplasm, cytoskeleton, microtubule organizing center, centrosome . Cytoplasm, cytoskeleton, microtubule organizing center, centrosome, centriole . Cytoplasm, cytoskeleton, spindle . Midbody . Chromosome . Perikaryon . Cell projection . Cell projection, growth cone . Myelin membrane . Localizes in the cytoplasm during most of the cell cycle except in the G2/M transition and during mitosis, where it is localized in association with chromatin and induces deacetylation of histone at 'Lys-16' (H4K16ac) (PubMed:17726514, PubMed:23468428). Colocalizes with KMT5A at mitotic foci (PubMed:23468428). Colocalizes with CDK1 at centrosome during prophase and splindle fibers during metaphase (PubMed:17488717). Colocalizes w
- Expression:
- Isoform 1 is expressed in heart, liver and skeletal muscle, weakly expressed in the cortex. Isoform 2 is strongly expressed in the cortex, weakly expressed in heart and liver. Weakly expressed in several malignancies including breast, liver, brain, kidney and prostate cancers compared to normal tissues. Weakly expressed in glioma cell lines compared to normal brain tissues (at protein level). Widely expressed. Highly expressed in heart, brain and skeletal muscle, while it is weakly expressed in placenta and lung. Down-regulated in many gliomas suggesting that it may act as a tumor suppressor gene in human gliomas possibly through the regulation of microtubule network.
CD90 affects the biological behavior and energy metabolism level of gastric cancer cells by targeting the PI3K/AKT/HIF‑1α signaling pathway. Oncology Letters Oncol Lett. 2021 Mar;21(3):1-1 WB Human 1:1000 AGS cell
- June 19-2018
- WESTERN IMMUNOBLOTTING PROTOCOL
- June 19-2018
- IMMUNOHISTOCHEMISTRY-PARAFFIN PROTOCOL
- June 19-2018
- IMMUNOFLUORESCENCE PROTOCOL
- September 08-2020
- FLOW-CYTOMEYRT-PROTOCOL
- May 20-2022
- Cell-Based ELISA│解您多样本WB检测之困扰
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-ACETYL-PROTEIN
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-PHOSPHO-PROTEIN
- July 13-2018
- Antibody-FAQs
- Products Images

- Western blot detection of SirT2 in PC-12, Rat Brain and Mouse Brain cell lysates using SirT2 mouse mAb (1:1000 diluted). Predicted band size: 39,43KDa. Observed band size:39,43KDa.

- Immunofluorescent analysis of Hela cells fixed fixed by anhydrous methanol at -20℃ and using SirT2 (200474,dilution 1:50) mouse mAb (green) and Histone H3.1 (Phospho-Ser10) (310045,dilution 1:200) Rabbit pAb (red).