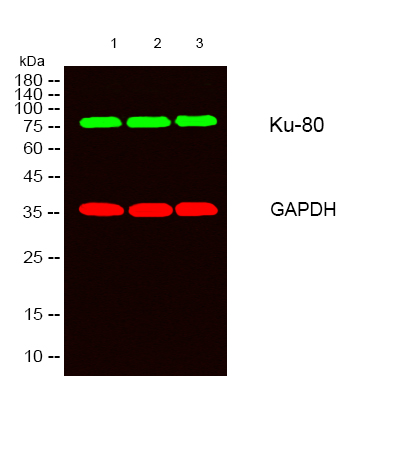
Total Ku70/80 Cell-Based Colorimetric ELISA Kit
- Catalog No.:KA4330C
- Applications:ELISA
- Reactivity:Human
- Gene Name:
- XRCC5
- Human Gene Id:
- 7520
- Human Swiss Prot No:
- P13010
- Mouse Swiss Prot No:
- P27641
- Storage Stability:
- 2-8°C/6 months
- Other Name:
- X-ray repair cross-complementing protein 5 (EC 3.6.4.-) (86 kDa subunit of Ku antigen) (ATP-dependent DNA helicase 2 subunit 2) (ATP-dependent DNA helicase II 80 kDa subunit) (CTC box-binding factor 85 kDa subunit) (CTC85) (CTCBF) (DNA repair protein XRCC5) (Ku80) (Ku86) (Lupus Ku autoantigen protein p86) (Nuclear factor IV) (Thyroid-lupus autoantigen) (TLAA) (X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining))
- Detection Method:
- Colorimetric
- Background:
- developmental stage:Expression increases during promyelocyte differentiation.,disease:Individuals with systemic lupus erythematosus (SLE) and related disorders produce extremely large amounts of autoantibodies to p70 and p86.,domain:The EEXXXDDL motif is required for the interaction with catalytic subunit PRKDC and its recruitment to sites of DNA damage.,function:Single stranded DNA-dependent ATP-dependent helicase. Has a role in chromosome translocation. The DNA helicase II complex binds preferentially to fork-like ends of double-stranded DNA in a cell cycle-dependent manner. It works in the 3'-5' direction. Binding to DNA may be mediated by p70. Involved in DNA nonhomologous end joining (NHEJ) required for double-strand break repair and V(D)J recombination. The Ku p70/p86 dimer acts as regulatory subunit of the DNA-dependent protein kinase complex DNA-PK by increasing the affinity of the catalytic subunit PRKDC to DNA by 100-fold. The Ku p70/p86 dimer is probably involved in stabilizing broken DNA ends and bringing them together. The assembly of the DNA-PK complex to DNA ends is required for the NHEJ ligation step. In association with NARG1, the Ku p70/p86 dimer binds to the osteocalcin promoter and activates osteocalcin expression.,induction:In osteoblasts, by FGF2.,PTM:Phosphorylated on serine residues. Phosphorylation by PRKDC may enhance helicase activity.,PTM:Sumoylated.,similarity:Belongs to the ku80 family.,similarity:Contains 1 Ku domain.,subunit:Heterodimer of a 70 kDa and a 80 kDa subunit. The dimer associates in a DNA-dependent manner with PRKDC to form the DNA-dependent protein kinase complex DNA-PK, and with the LIG4-XRCC4 complex. The dimer also associates with NARG1, and this complex displays DNA binding activity towards the osteocalcin FGF response element (OCFRE). In addition, the 80 kDa subunit binds to the osteoblast-specific transcription factors MSX2 and RUNX2. Interacts with ELF3. May interact with APLF.,
- Function:
- telomere maintenance, non-recombinational repair, immune system development, DNA metabolic process, DNA repair,double-strand break repair, double-strand break repair via nonhomologous end joining, DNA recombination, response to DNA damage stimulus, cell proliferation, positive regulation of cell development, regulation of cell death, DNA integration, viral reproduction, provirus integration, viral infectious cycle, initiation of viral infection, viral reproductive process, lysogeny, hemopoiesis, telomere organization, cellular response to stress, homeostatic process, regulation of apoptosis, negative regulation of apoptosis, regulation of programmed cell death, negative regulation of programmed cell death, positive regulation of cell differentiation, hemopoietic or lymphoid organ development, stem cell differentiation, regulation of neurogenesis, positive regulation of neurogenesis, posi
- Subcellular Location:
- Nucleus . Nucleus, nucleolus . Chromosome .
- June 19-2018
- WESTERN IMMUNOBLOTTING PROTOCOL
- June 19-2018
- IMMUNOHISTOCHEMISTRY-PARAFFIN PROTOCOL
- June 19-2018
- IMMUNOFLUORESCENCE PROTOCOL
- September 08-2020
- FLOW-CYTOMEYRT-PROTOCOL
- May 20-2022
- Cell-Based ELISA│解您多样本WB检测之困扰
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-ACETYL-PROTEIN
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-PHOSPHO-PROTEIN
- July 13-2018
- Antibody-FAQs