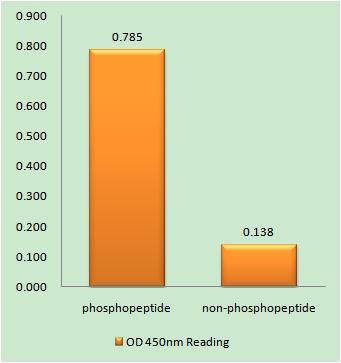
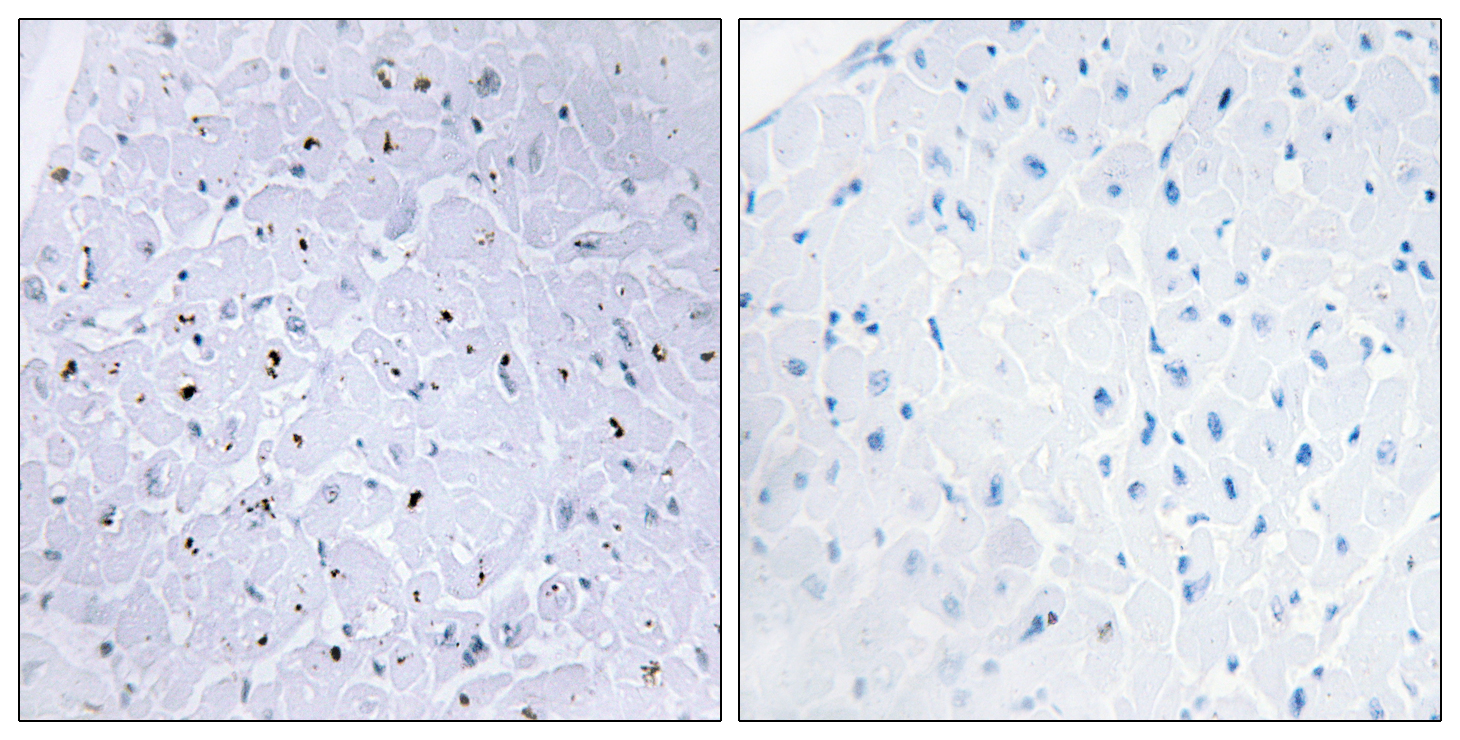
Total 53BP1 Cell-Based Colorimetric ELISA Kit
- Catalog No.:KA3981C
- Applications:ELISA
- Reactivity:Human;Mouse;Rat
- Gene Name:
- TP53BP1
- Human Gene Id:
- 7158
- Human Swiss Prot No:
- Q12888
- Mouse Swiss Prot No:
- P70399
- Storage Stability:
- 2-8°C/6 months
- Other Name:
- Tumor suppressor p53-binding protein 1 (53BP1) (p53-binding protein 1) (p53BP1)
- Detection Method:
- Colorimetric
- Background:
- function:May have a role in checkpoint signaling during mitosis (By similarity). Enhances TP53-mediated transcriptional activation. Plays a role in the response to DNA damage.,PTM:Asymmetrically dimethylated on Arg residues by PRMT1. Methylation is required for DNA binding.,PTM:Phosphorylated at basal level in the absence of DNA damage. Hyper-phosphorylated in an ATM-dependent manner in response to DNA damage induced by ionizing radiation. Hyper-phosphorylated in an ATR-dependent manner in response to DNA damage induced by UV irradiation.,similarity:Contains 2 BRCT domains.,subcellular location:Associated with kinetochores. Both nuclear and cytoplasmic in some cells. Recruited to sites of DNA damage, such as double stand breaks. Methylation of histone H4 at 'Lys-20' is required for efficient localization to double strand breaks.,subunit:Interacts with IFI202A (By similarity). Binds to the central domain of TP53/p53. May form homo-oligomers. Interacts with DCLRE1C. Interacts with histone H2AFX and this requires phosphorylation of H2AFX on 'Ser-139'. Interacts with histone H4 that has been dimethylated at 'Lys-20'. Has low affinity for histone H4 containing monomethylated 'Lys-20'. Does not bind histone H4 containing unmethylated or trimethylated 'Lys-20'. Has low affinity for histone H3 that has been dimethylated on 'Lys-79'. Has very low affinity for histone H3 that has been monomethylated on 'Lys-79' (in vitro). Does not bind unmethylated histone H3.,
- Function:
- DNA metabolic process, DNA repair, transcription, regulation of transcription, DNA-dependent, response to DNA damage stimulus, positive regulation of biosynthetic process, positive regulation of macromolecule biosynthetic process, positive regulation of macromolecule metabolic process, positive regulation of gene expression, positive regulation of cellular biosynthetic process, cellular response to stress, regulation of transcription, positive regulation of transcription, DNA-dependent, positive regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process, positive regulation of transcription, positive regulation of nitrogen compound metabolic process, regulation of RNA metabolic process, positive regulation of RNA metabolic process,
- Subcellular Location:
- Nucleus . Chromosome . Chromosome, centromere, kinetochore . Localizes to the nucleus in absence of DNA damage (PubMed:28241136). Following DNA damage, recruited to sites of DNA damage, such as double stand breaks (DSBs): recognizes and binds histone H2A monoubiquitinated at 'Lys-15' (H2AK15Ub) and histone H4 dimethylated at 'Lys-20' (H4K20me2), two histone marks that are present at DSBs sites (PubMed:23333306, PubMed:23760478, PubMed:24703952, PubMed:28241136, PubMed:17190600). Associated with kinetochores during mitosis (By similarity). .
- June 19-2018
- WESTERN IMMUNOBLOTTING PROTOCOL
- June 19-2018
- IMMUNOHISTOCHEMISTRY-PARAFFIN PROTOCOL
- June 19-2018
- IMMUNOFLUORESCENCE PROTOCOL
- September 08-2020
- FLOW-CYTOMEYRT-PROTOCOL
- May 20-2022
- Cell-Based ELISA│解您多样本WB检测之困扰
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-ACETYL-PROTEIN
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-PHOSPHO-PROTEIN
- July 13-2018
- Antibody-FAQs