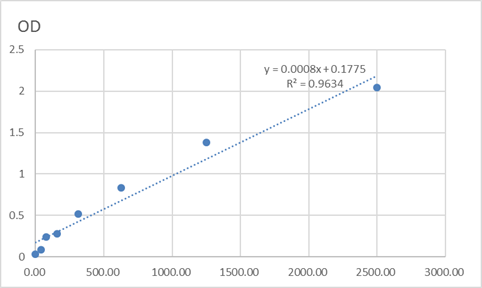
Total HBO1 Cell-Based Colorimetric ELISA Kit
- Catalog No.:KA3507C
- Applications:ELISA
- Reactivity:Human;Mouse;Rat
- Gene Name:
- MYST2
- Human Gene Id:
- 11143
- Human Swiss Prot No:
- O95251
- Mouse Swiss Prot No:
- Q5SVQ0
- Rat Swiss Prot No:
- Q810T5
- Storage Stability:
- 2-8°C/6 months
- Other Name:
- Histone acetyltransferase KAT7 (EC 2.3.1.48) (Histone acetyltransferase binding to ORC1) (Lysine acetyltransferase 7) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 2) (MYST-2)
- Detection Method:
- Colorimetric
- Background:
- catalytic activity:Acetyl-CoA + histone = CoA + acetylhistone.,domain:The C2HC-type zinc finger is required for interaction with MCM2 and ORC1L.,domain:The N-terminus is involved in transcriptional repression, while the C-terminus mediates AR-interaction.,function:Component of the HBO1 complex which has a histone H4-specific acetyltransferase activity, a reduced activity toward histone H3 and is responsible for the bulk of histone H4 acetylation in vivo. Through chromatin acetylation it may regulate DNA replication and act as a coactivator of TP53-dependent transcription. Specifically represses AR-mediated transcription.,PTM:Phosphorylated upon DNA damage, probably by ATM or ATR.,similarity:Belongs to the MYST (SAS/MOZ) family.,similarity:Contains 1 C2HC-type zinc finger.,subunit:Component of the HBO1 complex composed at least of ING4 or ING5, MYTS2/HBO1, EAF6, and one of PHF15, PHF16 and PHF17. Interacts with MCM2 and ORC1L. Interacts with the androgen receptor (AR) in the presence of dihydrotestosterone.,tissue specificity:Ubiquitously expressed, with highest levels in testis.,
- Function:
- DNA metabolic process, DNA replication, chromatin organization, transcription, regulation of transcription, DNA-dependent, protein amino acid acetylation, chromatin modification, covalent chromatin modification, histone modification, histone acetylation, protein amino acid acylation, histone H3 acetylation, histone H4 acetylation, histone H4-K5 acetylation, histone H4-K8 acetylation, histone H4-K12 acetylation, histone H4-K16 acetylation, regulation of transcription, regulation of RNA metabolic process, chromosome organization,
- Subcellular Location:
- Nucleus . Chromosome . Chromosome, centromere . Cytoplasm, cytosol . Associates with replication origins specifically during the G1 phase of the cell cycle (PubMed:18832067, PubMed:20129055). Localizes to transcription start sites (PubMed:21753189, PubMed:24065767). Localizes to ultraviolet-induced DNA damage sites following phosphorylation by ATR (PubMed:28719581). Localizes to centromeres in G1 phase (PubMed:27270040). .
- Expression:
- Ubiquitously expressed, with highest levels in testis.
- June 19-2018
- WESTERN IMMUNOBLOTTING PROTOCOL
- June 19-2018
- IMMUNOHISTOCHEMISTRY-PARAFFIN PROTOCOL
- June 19-2018
- IMMUNOFLUORESCENCE PROTOCOL
- September 08-2020
- FLOW-CYTOMEYRT-PROTOCOL
- May 20-2022
- Cell-Based ELISA│解您多样本WB检测之困扰
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-ACETYL-PROTEIN
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-PHOSPHO-PROTEIN
- July 13-2018
- Antibody-FAQs