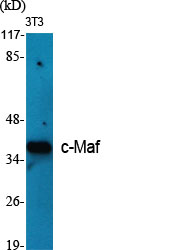
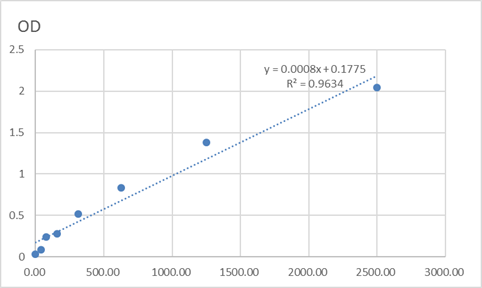
Total c-Maf Cell-Based Colorimetric ELISA Kit
- Catalog No.:KA3487C
- Applications:ELISA
- Reactivity:Human;Mouse;Rat
- Gene Name:
- MAF
- Human Gene Id:
- 4094
- Human Swiss Prot No:
- O75444
- Mouse Swiss Prot No:
- P54843
- Rat Swiss Prot No:
- P54844
- Storage Stability:
- 2-8°C/6 months
- Other Name:
- Transcription factor Maf (Proto-oncogene c-Maf) (V-maf musculoaponeurotic fibrosarcoma oncogene homolog)
- Detection Method:
- Colorimetric
- Background:
- disease:A chromosomal aberration involving MAF is found in some forms of multiple myeloma (MM). Translocation t(14;16)(q32.3;q23) with an IgH locus.,disease:Defects in MAF are the cause of congenital cerulean cataract 4 (CCA4) [MIM:610202]. CCA4 is a form of autosomal dominant congenital cataract (ADCC). Cerulean cataracts have peripheral bluish and white opacifications in concentric layers with occasional central lesions arranged radially. Although the opacities may be observed during fetal development and childhood, usually visual acuity is only mildly reduced until adulthood, when lens extraction is generally necessary.,disease:Defects in MAF are the cause of juvenile-onset pulverulent cataract [MIM:610202]. Cataract is a partial or complete ocular opacity that affects the crystalline lens or its capsule, leading to impaired vision or blindness.,function:Acts as a transcriptional activator or repressor. Involved in embryonic lens fiber cell development. Recruits the transcriptional coactivators CREBBP and/or EP300 to crystallin promoters leading to up-regulation of crystallin gene during lens fiber cell differentiation. Activates the expression of IL4 in T helper 2 (Th2) cells. Increases T cell susceptibility to apoptosis by interacting with MYB and decreasing BCL2 expression. Together with PAX6, transactivates strongly the glucagon gene promoter through the G1 element. Activates transcription of the CD13 proximal promoter in endothelial cells. Represses transcription of the CD13 promoter in early stages of myelopoiesis by affecting the ETS1 and MYB cooperative interaction. Involved in the initial chondrocyte terminal differentiation and the disappearance of hypertrophic chondrocytes during endochondral bone development. Binds to the sequence 5'-[GT]G[GC]N[GT]NCTCAGNN-3' in the L7 promoter. Binds to the T-MARE (Maf response element) sites of lens-specific alpha- and beta-crystallin gene promoters. Binds element G1 on the glucagon promoter. Binds an AT-rich region adjacent to the TGC motif (atypical Maf response element) in the CD13 proximal promoter in endothelial cells (By similarity). When overexpressed, represses anti-oxidant reponse element (ARE)-mediated transcription. Involved either as an oncogene or as a tumor suppressor, depending on the cell context. Binds to the ARE sites of detoxifying enzyme gene promoters.,induction:Up-regulated with tert-butyl hydroquinone (t-BHQ).,PTM:Phosphorylated by GSK3 and MAPK13 on serine and threonine residues (Probable). The posphorylation status can serve to either stimulate or inhibit transcription.,PTM:Ubiquitinated, leading to its degradation by the proteasome. Ubiquitination is triggered by glucocorticoids.,similarity:Belongs to the bZIP family.,similarity:Belongs to the bZIP family. Maf subfamily.,similarity:Contains 1 bZIP domain.,subunit:Homodimer or heterodimer with other bHLH-Zip transcription factors. Binds DNA as a homodimer or as a heterodimer. Heterotetramer of two MAF and two USF2. Interacts with PAX6; the interaction is direct. Interacts with MYB; interaction take place weakly in normal T cells and increases in T cells following stimulation through the TCR engagement. Interacts with MYB; the ternary complexe formed with MYB and the CD13 promoter is regulated in response to differentiating signals. Interacts with USF2; the interaction inhibits its DNA-binding activity on the L7 promoter. Interacts with CREBBP, EP300 and ETS1.,tissue specificity:Expressed in endothelial cells.,
- Function:
- eye development, cytokine production, lens development in camera-type eye, transcription, transcription, DNA-dependent, regulation of transcription, DNA-dependent, regulation of transcription from RNA polymerase II promoter,transcription from RNA polymerase II promoter, sensory organ development, positive regulation of biosynthetic process, positive regulation of macromolecule biosynthetic process, positive regulation of macromolecule metabolic process, positive regulation of gene expression, positive regulation of cellular biosynthetic process, regulation of chondrocyte differentiation, RNA biosynthetic process, camera-type eye development, regulation of transcription,positive regulation of transcription, DNA-dependent, positive regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process, positive regulation of transcription, positive regulation of transcription
- Subcellular Location:
- Nucleus .
- Expression:
- Expressed in endothelial cells.
- June 19-2018
- WESTERN IMMUNOBLOTTING PROTOCOL
- June 19-2018
- IMMUNOHISTOCHEMISTRY-PARAFFIN PROTOCOL
- June 19-2018
- IMMUNOFLUORESCENCE PROTOCOL
- September 08-2020
- FLOW-CYTOMEYRT-PROTOCOL
- May 20-2022
- Cell-Based ELISA│解您多样本WB检测之困扰
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-ACETYL-PROTEIN
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-PHOSPHO-PROTEIN
- July 13-2018
- Antibody-FAQs