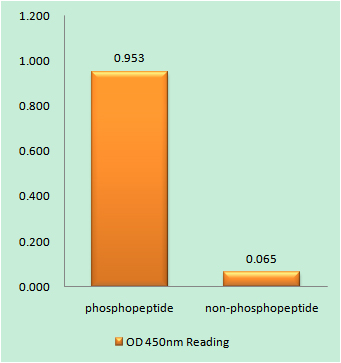
Total Hrs Cell-Based Colorimetric ELISA Kit
- Catalog No.:KA3453C
- Applications:ELISA
- Reactivity:Human;Mouse;Rat
- Gene Name:
- HGS
- Human Gene Id:
- 9146
- Human Swiss Prot No:
- O14964
- Mouse Swiss Prot No:
- Q99LI8
- Rat Swiss Prot No:
- Q9JJ50
- Storage Stability:
- 2-8°C/6 months
- Other Name:
- Hepatocyte growth factor-regulated tyrosine kinase substrate (Hrs) (Protein pp110)
- Detection Method:
- Colorimetric
- Background:
- domain:Has a double-sided UIM that can bind 2 ubiquitin molecules, one on each side of the helix.,function:Involved in intracellular signal transduction mediated by cytokines and growth factors. When associated with STAM, it suppresses DNA signaling upon stimulation by IL-2 and GM-CSF. Could be a direct effector of PI3-kinase in vesicular pathway via early endosomes and may regulate trafficking to early and late endosomes by recruiting clathrin. May concentrate ubiquitinated receptors within clathrin-coated regions. Involved in down-regulation of receptor tyrosine kinase via multivesicular body (MVBs) when complexed with STAM (ESCRT-0 complex). The ESCRT-0 complex binds ubiquitin and acts as sorting machinery that recognizes ubiquitinated receptors and transfers them to further sequential lysosomal sorting/trafficking processes. May contribute to the efficient recruitment of SMADs to the activin receptor complex.,PTM:Phosphorylated on Tyr-334. A minor site of phosphorylation on Tyr-329 is detected (By similarity). Phosphorylation occurs in response to EGF, IL-2, GM-CSF and HGF.,similarity:Contains 1 FYVE-type zinc finger.,similarity:Contains 1 UIM (ubiquitin-interacting motif) repeat.,similarity:Contains 1 VHS domain.,subunit:Component of the ESCRT-0 complex composed of STAM or STAM2 and HGS. Part of a complex at least composed of HSG, STAM2 (or probably STAM) and EPS15. Interacts with STAM. Interacts with STAM2. Interacts with EPS15; the interaction is direct, calcium-dependent and inhibited by SNAP25. Interacts with NF2; the interaction is direct. Interacts with ubiquitin; the interaction is direct. Interacts with VPS37C. Interacts with SMAD1, SMAD2 and SMAD3. Interacts with TSG101; the interaction mediates the association with the ESCRT-I complex. Interacts with SNAP25; the interaction is direct and decreases with addition of increasing concentrations of free calcium. Interacts with SNX1; the interaction is direct. Component of a 550 kDa membrane complex at least composed of HGS and SNX1 but excluding EGFR.,tissue specificity:Ubiquitous expression in adult and fetal tissues with higher expression in testis and peripheral blood leukocytes.,
- Function:
- intracellular protein transport, protein localization, negative regulation of cell proliferation, regulation of catabolic process, negative regulation of signal transduction, regulation of protein kinase cascade, negative regulation of cell communication, negative regulation of protein kinase cascade, protein transport, vesicle-mediated transport,endosome transport, cellular protein localization, regulation of cell proliferation, regulation of protein catabolic process, establishment of protein localization, regulation of JAK-STAT cascade, negative regulation of JAK-STAT cascade, intracellular transport, cellular macromolecule localization,
- Subcellular Location:
- Cytoplasm . Early endosome membrane ; Peripheral membrane protein ; Cytoplasmic side . Endosome, multivesicular body membrane ; Peripheral membrane protein . Colocalizes with UBQLN1 in ubiquitin-rich cytoplasmic aggregates that are not endocytic compartments. .
- Expression:
- Ubiquitous expression in adult and fetal tissues with higher expression in testis and peripheral blood leukocytes.
- June 19-2018
- WESTERN IMMUNOBLOTTING PROTOCOL
- June 19-2018
- IMMUNOHISTOCHEMISTRY-PARAFFIN PROTOCOL
- June 19-2018
- IMMUNOFLUORESCENCE PROTOCOL
- September 08-2020
- FLOW-CYTOMEYRT-PROTOCOL
- May 20-2022
- Cell-Based ELISA│解您多样本WB检测之困扰
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-ACETYL-PROTEIN
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-PHOSPHO-PROTEIN
- July 13-2018
- Antibody-FAQs