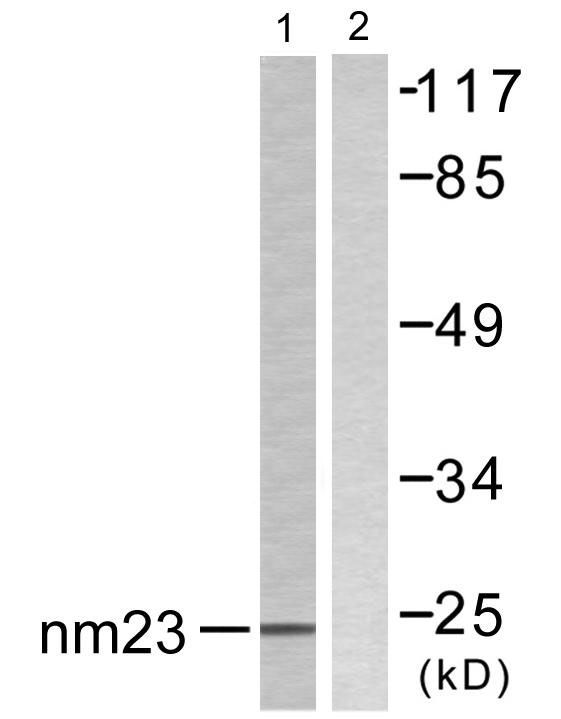
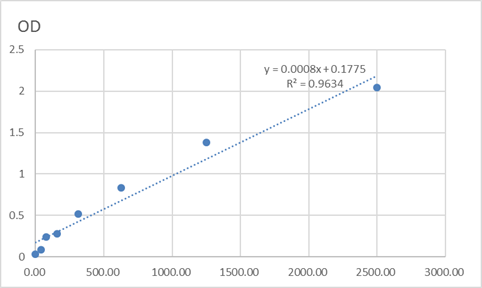
Total NM23-H2 Cell-Based Colorimetric ELISA Kit
- Catalog No.:KA3338C
- Applications:ELISA
- Reactivity:Human;Mouse;Rat
- Gene Name:
- NME2
- Human Gene Id:
- 4831
- Human Swiss Prot No:
- P22392
- Mouse Swiss Prot No:
- Q01768
- Rat Swiss Prot No:
- P19804
- Storage Stability:
- 2-8°C/6 months
- Other Name:
- Nucleoside diphosphate kinase B (NDK B) (NDP kinase B) (EC 2.7.4.6) (C-myc purine-binding transcription factor PUF) (Histidine protein kinase NDKB) (EC 2.7.13.3) (nm23-H2)
- Detection Method:
- Colorimetric
- Background:
- catalytic activity:ATP + nucleoside diphosphate = ADP + nucleoside triphosphate.,cofactor:Magnesium.,disease:This protein is found in reduced amount in tumor cells of high metastatic potential.,disease:This protein is found in reduced amount in tumor cells of high metastatic potential. Somatic mutations of NME1 are found in neuroblastoma. Increased NME1 in neuroblastoma is correlated with features of the disease that are associated with aggressive tumors. May therefore have distinct if not opposite roles in different tumors.,enzyme regulation:Autophosphorylation at His-118 increases serine/threonine protein kinase activity of the enzyme. Interaction with the SET complex inhibits exonuclease activity.,function:Major role in the synthesis of nucleoside triphosphates other than ATP. Negatively regulates Rho activity by interacting with AKAP13/LBC. Acts as a transcriptional activator of the c-Myc gene; binds DNA non-specifically (PubMed:8392752).,function:Major role in the synthesis of nucleoside triphosphates other than ATP. Possesses nucleoside-diphosphate kinase, serine/threonine-specific protein kinase, geranyl and farnesyl pyrophosphate kinase, histidine protein kinase and 3'-5' exonuclease activities. Involved in cell proliferation, differentiation and development, signal transduction, G protein-coupled receptor endocytosis, and gene expression. Required for neural development including neural patterning and cell fate determination. Has tumor metastasis-suppressive capacity.,PTM:The N-terminus is blocked.,similarity:Belongs to the NDK family.,subcellular location:Cell-cycle dependent nuclear localization which can be induced by interaction with Epstein-barr viral proteins or by degradation of the SET complex by GzmA.,subcellular location:Isoform 2 is mainly cytoplasmic and isoform 1 and isoform 2 are excluded from the nucleolus.,subunit:Hexamer of two different chains: A and B (A6, A5B, A4B2, A3B3, A2B4, AB5, B6). Interacts with CAPN8 (By similarity). Interacts with AKAP13.,subunit:Hexamer of two different chains: A and B (A6, A5B, A4B2, A3B3, A2B4, AB5, B6). Interacts with SET and PRUNE.,tissue specificity:Isoform 1 is expressed in heart, brain, placenta, lung, liver, skeletal muscle, pancreas, spleen and thymus. Expressed in lung carcinoma cell lines but not in normal lung tissues. Isoform 2 is ubiquitously expressed and its expression is also related to tumor differentiation. Isoform 3 is ubiquitously expressed.,tissue specificity:Ubiquitously expressed.,
- Function:
- regulation of myeloid leukocyte differentiation, negative regulation of myeloid leukocyte differentiation, purine nucleotide metabolic process, purine nucleotide biosynthetic process, nucleoside diphosphate phosphorylation, GTP biosynthetic process, pyrimidine nucleoside metabolic process, pyrimidine nucleotide metabolic process, pyrimidine nucleotide biosynthetic process, UTP biosynthetic process, CTP biosynthetic process, transcription, regulation of transcription, DNA-dependent, phosphorus metabolic process, phosphate metabolic process, endocytosis, cell adhesion, lactation, positive regulation of cell proliferation, negative regulation of cell proliferation, nucleoside metabolic process, ribonucleoside metabolic process, nucleoside diphosphate metabolic process, nucleoside triphosphate metabolic process, nucleoside triphosphate biosynthetic process, purine nucleoside triphosphate met
- Subcellular Location:
- Cytoplasm . Cell projection, lamellipodium . Cell projection, ruffle . Colocalizes with ITGB1 and ITGB1BP1 at the edge or peripheral ruffles and lamellipodia during the early stages of cell spreading on fibronectin or collagen but not on vitronectin or laminin substrates. .; [Isoform 1]: Cytoplasm . Cytoplasm, perinuclear region . Nucleus .; [Isoform 3]: Cytoplasm . Cytoplasm, perinuclear region . Nucleus .
- Expression:
- [Isoform 1]: Ubiquitously expressed. ; [Isoform 3]: Ubiquitously expressed.
- June 19-2018
- WESTERN IMMUNOBLOTTING PROTOCOL
- June 19-2018
- IMMUNOHISTOCHEMISTRY-PARAFFIN PROTOCOL
- June 19-2018
- IMMUNOFLUORESCENCE PROTOCOL
- September 08-2020
- FLOW-CYTOMEYRT-PROTOCOL
- May 20-2022
- Cell-Based ELISA│解您多样本WB检测之困扰
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-ACETYL-PROTEIN
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-PHOSPHO-PROTEIN
- July 13-2018
- Antibody-FAQs