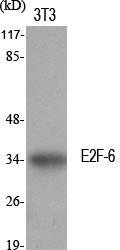
Total E2F-6 Cell-Based Colorimetric ELISA Kit
- Catalog No.:KA3248C
- Applications:ELISA
- Reactivity:Human;Mouse;Rat
- Gene Name:
- E2F6
- Human Gene Id:
- 1876
- Human Swiss Prot No:
- O75461
- Mouse Swiss Prot No:
- O54917
- Storage Stability:
- 2-8°C/6 months
- Other Name:
- Transcription factor E2F6 (E2F-6)
- Detection Method:
- Colorimetric
- Background:
- function:Inhibitor of E2F-dependent transcription. Binds DNA cooperatively with DP proteins through the E2 recognition site, 5'-TTTC[CG]CGC-3'. Has a preference for the 5'-TTTCCCGC-3' E2F recognition site. E2F-6 lacks the transcriptional activation and pocket protein binding domains. Appears to regulate a subset of E2F-dependent genes whose products are required for entry into the cell cycle but not for normal cell cycle progression. May silence expression via the recruitment of a chromatin remodeling complex containing histone H3-K9 methyltransferase activity. Overexpression delays the exit of cells from the S-phase.,similarity:Belongs to the E2F/DP family.,subunit:Component of the DRTF1/E2F transcription factor complex. Forms heterodimers with DP family members. Part of the E2F6.com-1 complex in G0 phase composed of E2F6, MGA, MAX, TFDP1, CBX3, BAT8, EUHMTASE1, RING1, RNF2, MBLR, L3MBTL2 and YAF2.,tissue specificity:Expressed in all tissues examined. Highest levels in placenta, skeletal muscle, heart, ovary, kidney, small intestine and spleen.,
- Function:
- G1/S transition of mitotic cell cycle, regulation of transcription of G1/S-phase of mitotic cell cycle, negative regulation of transcription from RNA polymerase II promoter, mitotic cell cycle, transcription, regulation of transcription, DNA-dependent, regulation of transcription from RNA polymerase II promoter, cell cycle, negative regulation of biosynthetic process, negative regulation of macromolecule biosynthetic process, negative regulation of macromolecule metabolic process, negative regulation of gene expression, negative regulation of transcription, cell cycle process, cell cycle phase, negative regulation of cellular biosynthetic process, regulation of transcription, negative regulation of transcription, DNA-dependent, negative regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process, negative regulation of nitrogen compound metabolic process, regulati
- Subcellular Location:
- Nucleus .
- Expression:
- Expressed in all tissues examined. Highest levels in placenta, skeletal muscle, heart, ovary, kidney, small intestine and spleen.
- June 19-2018
- WESTERN IMMUNOBLOTTING PROTOCOL
- June 19-2018
- IMMUNOHISTOCHEMISTRY-PARAFFIN PROTOCOL
- June 19-2018
- IMMUNOFLUORESCENCE PROTOCOL
- September 08-2020
- FLOW-CYTOMEYRT-PROTOCOL
- May 20-2022
- Cell-Based ELISA│解您多样本WB检测之困扰
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-ACETYL-PROTEIN
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-PHOSPHO-PROTEIN
- July 13-2018
- Antibody-FAQs