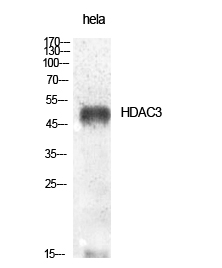
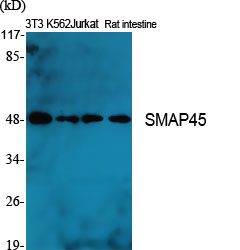
Phospho HDAC3 (S424) Cell-Based Colorimetric ELISA Kit
- Catalog No.:KA1463C
- Applications:ELISA
- Reactivity:Human;Mouse;Rat
- Gene Name:
- HDAC3
- Human Gene Id:
- 8841
- Human Swiss Prot No:
- O15379
- Mouse Swiss Prot No:
- O88895
- Rat Swiss Prot No:
- Q6P6W3
- Storage Stability:
- 2-8°C/6 months
- Other Name:
- Histone deacetylase 3 (HD3) (EC 3.5.1.98) (RPD3-2) (SMAP45)
- Detection Method:
- Colorimetric
- Background:
- catalytic activity:Hydrolysis of an N(6)-acetyl-lysine residue of a histone to yield a deacetylated histone.,function:Responsible for the deacetylation of lysine residues on the N-terminal part of the core histones (H2A, H2B, H3 and H4). Histone deacetylation gives a tag for epigenetic repression and plays an important role in transcriptional regulation, cell cycle progression and developmental events. Histone deacetylases act via the formation of large multiprotein complexes. Probably participates in the regulation of transcription through its binding to the zinc-finger transcription factor YY1; increases YY1 repression activity. Required to repress transcription of the POU1F1 transcription factor.,PTM:Sumoylated in vitro.,similarity:Belongs to the histone deacetylase family. Type 1 subfamily.,subunit:Interacts with HDAC7 and HDAC9. Forms a heterologous complex at least with YY1. Interacts with DAXX, HDAC10 and DACH1. Found in a complex with NCOR1 and NCOR2. Component of the N-Cor repressor complex, at least composed of NCOR1, NCOR2, HDAC3, TBL1X, TBL1R, CORO2A and GPS2. Interacts with BCOR, MJD2A/JHDM3A, NRIP1, PRDM6 and SRY. Interacts with BTBD14B. Interacts with GLIS2 (By similarity). Interacts with CBFA2T3.,tissue specificity:Widely expressed.,
- Function:
- negative regulation of transcription from RNA polymerase II promoter, microtubule cytoskeleton organization, M phase, angiogenesis, blood vessel development, liver development, vasculature development, chromatin organization,transcription, regulation of transcription, DNA-dependent, regulation of transcription from RNA polymerase II promoter,protein amino acid deacetylation, anti-apoptosis, cytoskeleton organization, microtubule-based process, cell cycle,spindle organization, regulation of mitotic cell cycle, negative regulation of biosynthetic process, negative regulation of signal transduction, regulation of specific transcription from RNA polymerase II promoter, negative regulation of specific transcription from RNA polymerase II promoter, negative regulation of macromolecule biosynthetic process,negative regulation of macromolecule metabolic process, regulation of protein kinase casc
- Subcellular Location:
- Nucleus . Cytoplasm . Cytoplasm, cytosol . Colocalizes with XBP1 and AKT1 in the cytoplasm (PubMed:25190803). Predominantly expressed in the nucleus in the presence of CCAR2 (PubMed:21030595). .
- Expression:
- Widely expressed.
- June 19-2018
- WESTERN IMMUNOBLOTTING PROTOCOL
- June 19-2018
- IMMUNOHISTOCHEMISTRY-PARAFFIN PROTOCOL
- June 19-2018
- IMMUNOFLUORESCENCE PROTOCOL
- September 08-2020
- FLOW-CYTOMEYRT-PROTOCOL
- May 20-2022
- Cell-Based ELISA│解您多样本WB检测之困扰
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-ACETYL-PROTEIN
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-PHOSPHO-PROTEIN
- July 13-2018
- Antibody-FAQs