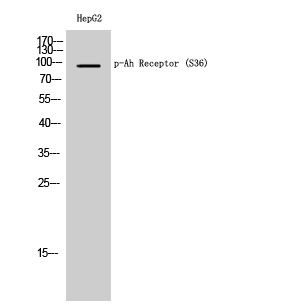
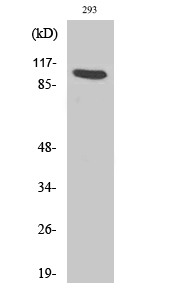
Phospho AhR (S36) Cell-Based Colorimetric ELISA Kit
- Catalog No.:KA1395C
- Applications:ELISA
- Reactivity:Human;Mouse;Rat
- Gene Name:
- AHR
- Human Swiss Prot No:
- P35869/A9YTQ3
- Storage Stability:
- 2-8°C/6 months
- Detection Method:
- Colorimetric
- Background:
- function:Ligand-activated transcriptional activator. Binds to the XRE promoter region of genes it activates. Activates the expression of multiple phase I and II xenobiotic chemical metabolizing enzyme genes (such as the CYP1A1 gene). Mediates biochemical and toxic effects of halogenated aromatic hydrocarbons. Involved in cell-cycle regulation. Likely to play an important role in the development and maturation of many tissues.,induction:Induced or repressed by TGF-beta and dioxin in a cell-type specific fashion. Repressed by cAMP, retinoic acid, and TPA.,similarity:Contains 1 basic helix-loop-helix (bHLH) domain.,similarity:Contains 1 PAC (PAS-associated C-terminal) domain.,similarity:Contains 2 PAS (PER-ARNT-SIM) domains.,subcellular location:Initially cytoplasmic; upon binding with ligand and interaction with a HSP90, it translocates to the nucleus.,subunit:Binds MYBBP1A (By similarity). Efficient DNA binding requires dimerization with another bHLH protein. In the nucleus, heterodimer of AHR and ARNT. Interacts with coactivators including SRC-1, RIP140 and NOCA7, and with the corepressor SMRT. Interacts with NEDD8 and IVNS1ABP.,tissue specificity:Expressed in all tissues tested including blood, brain, heart, kidney, liver, lung, pancreas and skeletal muscle.,
- Function:
- transcription, transcription, DNA-dependent, regulation of transcription, DNA-dependent, regulation of transcription from RNA polymerase II promoter, transcription from RNA polymerase II promoter, apoptosis, cell cycle, cell death,response to xenobiotic stimulus, positive regulation of biosynthetic process, positive regulation of macromolecule biosynthetic process, positive regulation of macromolecule metabolic process, positive regulation of gene expression,programmed cell death, death, positive regulation of cellular biosynthetic process, RNA biosynthetic process,regulation of protein complex assembly, regulation of cellular component biogenesis, regulation of transcription,positive regulation of transcription, DNA-dependent, regulation of transcriptional preinitiation complex assembly,positive regulation of transcriptional preinitiation complex assembly, positive regulation of nucleob
- Subcellular Location:
- Cytoplasm . Nucleus . Initially cytoplasmic; upon binding with ligand and interaction with a HSP90, it translocates to the nucleus. .
- Expression:
- Expressed in all tissues tested including blood, brain, heart, kidney, liver, lung, pancreas and skeletal muscle. Expressed in retinal photoreceptors (PubMed:29726989).
- June 19-2018
- WESTERN IMMUNOBLOTTING PROTOCOL
- June 19-2018
- IMMUNOHISTOCHEMISTRY-PARAFFIN PROTOCOL
- June 19-2018
- IMMUNOFLUORESCENCE PROTOCOL
- September 08-2020
- FLOW-CYTOMEYRT-PROTOCOL
- May 20-2022
- Cell-Based ELISA│解您多样本WB检测之困扰
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-ACETYL-PROTEIN
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-PHOSPHO-PROTEIN
- July 13-2018
- Antibody-FAQs