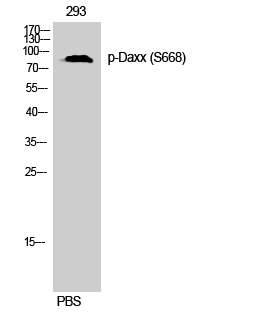
Phospho Daxx (S668) Cell-Based Colorimetric ELISA Kit
- Catalog No.:KA1244C
- Applications:ELISA
- Reactivity:Human
- Gene Name:
- DAXX
- Human Gene Id:
- 1616
- Human Swiss Prot No:
- Q9UER7
- Mouse Swiss Prot No:
- O35613
- Storage Stability:
- 2-8°C/6 months
- Other Name:
- Death domain-associated protein 6 (Daxx) (hDaxx) (ETS1-associated protein 1) (EAP1) (Fas death domain-associated protein)
- Detection Method:
- Colorimetric
- Background:
- function:Proposed to mediate activation of the JNK pathway and apoptosis via MAP3K5 in response to signaling from TNFRSF6 and TGFBR2. Interaction with HSPB1/HSP27 may prevent interaction with TNFRSF6 and MAP3K5 and block DAXX-mediated apoptosis. In contrast, in lymphoid cells JNC activation and TNFRSF6-mediated apoptosis may not involve DAXX. Seems to regulate transcription in PML/POD/ND10 nuclear bodies together with PML and may influence TNFRSF6-dependent apoptosis thereby. Down-regulates basal and activated transcription. Seems to act as a transcriptional co-repressor and inhibits PAX3 and ETS1 through direct protein-protein interaction. Modulates PAX5 activity. Its transcription repressor activity is modulated by recruiting it to subnuclear compartments like the nucleolus or PML/POD/ND10 nuclear bodies through interactions with MCSR1 and PML, respectively.,induction:Upon mitogenic stimulation by concanavalin A.,PTM:Phosphorylated upon DNA damage, probably by ATM or ATR. Phosphorylated by HIPK1 upon glucose deprivation.,PTM:Polyubiquitinated; which is promoted by CUL3 and SPOP and results in proteasomal degradation.,PTM:Sumoylated.,similarity:Belongs to the DAXX family.,subcellular location:Dispersed throughout the nucleoplasm, in PML/POD/ND10 nuclear bodies, and in nucleoli. Colocalizes with a subset of interphase centromeres, but is absent from mitotic centromeres. Detected in cytoplasmic punctate structures. Translocates from the nucleus to the cytoplasm upon glucose deprivation or oxidative stress.,subunit:Homomultimer. Binds to the TNFRSF6 death domain via its C-terminus and to PAX5. Binds to SLC2A4/GLUT4, MAP3K5, TGFBR2, phosphorylated dimeric HSPB1/HSP27, CENPC1, ETS1, sumoylated PML, UBE2I and MCRS1. Is part of a complex containing PAX5 and CREBBP. Interacts with HIPK2 and HIPK3 via its N-terminus. Interacts with HIPK1, which induces translocation from PML/POD/ND10 nuclear bodies to chromatin and enhances association with HDAC1 (By similarity). The non-phosphorylated form binds to PAX3, PAX7, DEK, HDAC1, HDAC2, HDAC3, acetylated histone H4 and histones H2A, H2B, H3 and H4. Interacts with SPOP. Part of a complex consisting of DAXX, CUL3 and SPOP. Interacts with CBP; the interaction is dependent the sumoylation of CBP and suppresses CBP transcriptional activity via recruitment of HDAC2 (By similarity). Interacts with HCMV tegument phosphoprotein pp71.,tissue specificity:Ubiquitous.,
- Function:
- MAPKKK cascade, activation of MAPK activity, cytokinesis after mitosis, cytokinesis, regulation of protein amino acid phosphorylation, transcription, regulation of transcription, DNA-dependent, protein amino acid phosphorylation,phosphorus metabolic process, phosphate metabolic process, apoptosis, induction of apoptosis, cell cycle, intracellular signaling cascade, protein kinase cascade, JNK cascade, activation of JUN kinase activity, cell death, induction of apoptosis by extracellular signals, negative regulation of biosynthetic process, negative regulation of macromolecule biosynthetic process, negative regulation of macromolecule metabolic process, regulation of protein kinase cascade,negative regulation of gene expression, regulation of cell death, positive regulation of cell death, programmed cell death, induction of programmed cell death, death, phosphorylation, negative regulatio
- Subcellular Location:
- Cytoplasm . Nucleus, nucleoplasm . Nucleus, PML body . Nucleus, nucleolus . Chromosome, centromere . Dispersed throughout the nucleoplasm, in PML/POD/ND10 nuclear bodies, and in nucleoli (Probable). Colocalizes with histone H3.3, ATRX, HIRA and ASF1A at PML-nuclear bodies (PubMed:12953102, PubMed:14990586, PubMed:23222847, PubMed:24200965). Colocalizes with a subset of interphase centromeres, but is absent from mitotic centromeres (PubMed:9645950). Detected in cytoplasmic punctate structures (PubMed:11842083). Translocates from the nucleus to the cytoplasm upon glucose deprivation or oxidative stress (PubMed:12968034). Colocalizes with RASSF1 in the nucleus (PubMed:18566590). Colocalizes with USP7 in nucleoplasma with accumulation in speckled structures (PubMed:16845383). .; [Isoform beta]
- Expression:
- Ubiquitous.
- June 19-2018
- WESTERN IMMUNOBLOTTING PROTOCOL
- June 19-2018
- IMMUNOHISTOCHEMISTRY-PARAFFIN PROTOCOL
- June 19-2018
- IMMUNOFLUORESCENCE PROTOCOL
- September 08-2020
- FLOW-CYTOMEYRT-PROTOCOL
- May 20-2022
- Cell-Based ELISA│解您多样本WB检测之困扰
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-ACETYL-PROTEIN
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-PHOSPHO-PROTEIN
- July 13-2018
- Antibody-FAQs
.jpg)