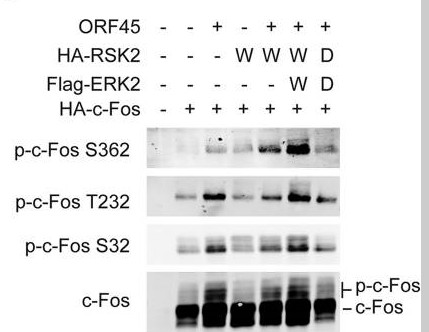
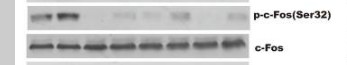
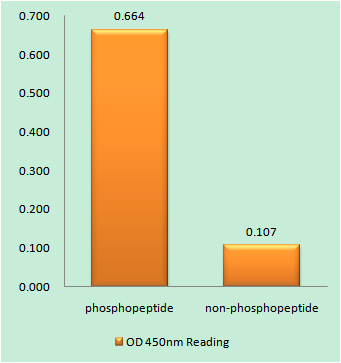
Phospho FOS (S32) Cell-Based Colorimetric ELISA Kit
- Catalog No.:KA1726C
- Applications:ELISA
- Reactivity:Human;Mouse;Rat
- Gene Name:
- FOS
- Human Gene Id:
- 2353
- Human Swiss Prot No:
- P01100
- Mouse Swiss Prot No:
- P01101
- Rat Swiss Prot No:
- P12841
- Storage Stability:
- 2-8°C/6 months
- Other Name:
- Proto-oncogene c-Fos (Cellular oncogene fos) (G0/G1 switch regulatory protein 7)
- Detection Method:
- Colorimetric
- Background:
- function:Nuclear phosphoprotein which forms a tight but non-covalently linked complex with the JUN/AP-1 transcription factor. In the heterodimer, c-fos and JUN/AP-1 basic regions each seems to interact with symmetrical DNA half sites. Has a critical function in regulating the development of cells destined to form and maintain the skeleton. It is thought to have an important role in signal transduction, cell proliferation and differentiation.,PTM:Constitutively sumoylated by SUMO1, SUMO2 and SUMO3. Desumoylated by SENP2. Sumoylation requires heterodimerization with JUN and is enhanced by mitogen stimulation. Sumoylation inhibits the AP-1 transcriptional activity and is, itself, inhibited by Ras-activated phosphorylation on Thr-232.,PTM:Phosphorylated in the C-terminal upon stimulation by nerve growth factor (NGF) and epidermal growth factor (EGF). Phosphorylated, in vitro, by MAPK and RSK1. Phosphorylation on both Ser-362 and Ser-374 by MAPK1/2 and RSK1/2 leads to protein stabilization with phosphorylation on Ser-374 being the major site for protein stabilization on NGF stimulation. Phosphorylation on Ser-362 and Ser-374 primes further phosphorylations on Thr-325 and Thr-331 through promoting docking of MAPK to the DEF domain. Phosphorylation on Thr-232, induced by HA-RAS, activates the transcriptional activity and antagonizes sumoylation. Phosphorylation on Ser-362 by RSK2 in osteoblasts contributes to osteoblast transformation.,similarity:Belongs to the bZIP family.,similarity:Belongs to the bZIP family. Fos subfamily.,similarity:Contains 1 bZIP domain.,subunit:Heterodimer with JUN. Interacts with DSIPI; this interaction inhibits the binding of active AP1 to its target DNA. Interacts with MAFB.,
- Function:
- response to reactive oxygen species, conditioned taste aversion, response to molecule of bacterial origin, DNA metabolic process, DNA modification, DNA alkylation, DNA methylation, regulation of transcription, DNA-dependent,regulation of transcription from RNA polymerase II promoter, one-carbon metabolic process, defense response,inflammatory response, response to oxidative stress, cell surface receptor linked signal transduction, enzyme linked receptor protein signaling pathway, transmembrane receptor protein serine/threonine kinase signaling pathway,transforming growth factor beta receptor signaling pathway, female pregnancy, aging, behavior, learning or memory,learning, feeding behavior, associative learning, response to temperature stimulus, response to radiation, response to cold, response to light stimulus, response to wounding, response to mechanical stimulus, response to bacteriu
- Subcellular Location:
- Nucleus. Endoplasmic reticulum. Cytoplasm, cytosol. In quiescent cells, present in very small amounts in the cytosol. Following induction of cell growth, first localizes to the endoplasmic reticulum and only later to the nucleus. Localization at the endoplasmic reticulum requires dephosphorylation at Tyr-10 and Tyr-30.
- June 19-2018
- WESTERN IMMUNOBLOTTING PROTOCOL
- June 19-2018
- IMMUNOHISTOCHEMISTRY-PARAFFIN PROTOCOL
- June 19-2018
- IMMUNOFLUORESCENCE PROTOCOL
- September 08-2020
- FLOW-CYTOMEYRT-PROTOCOL
- May 20-2022
- Cell-Based ELISA│解您多样本WB检测之困扰
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-ACETYL-PROTEIN
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-PHOSPHO-PROTEIN
- July 13-2018
- Antibody-FAQs