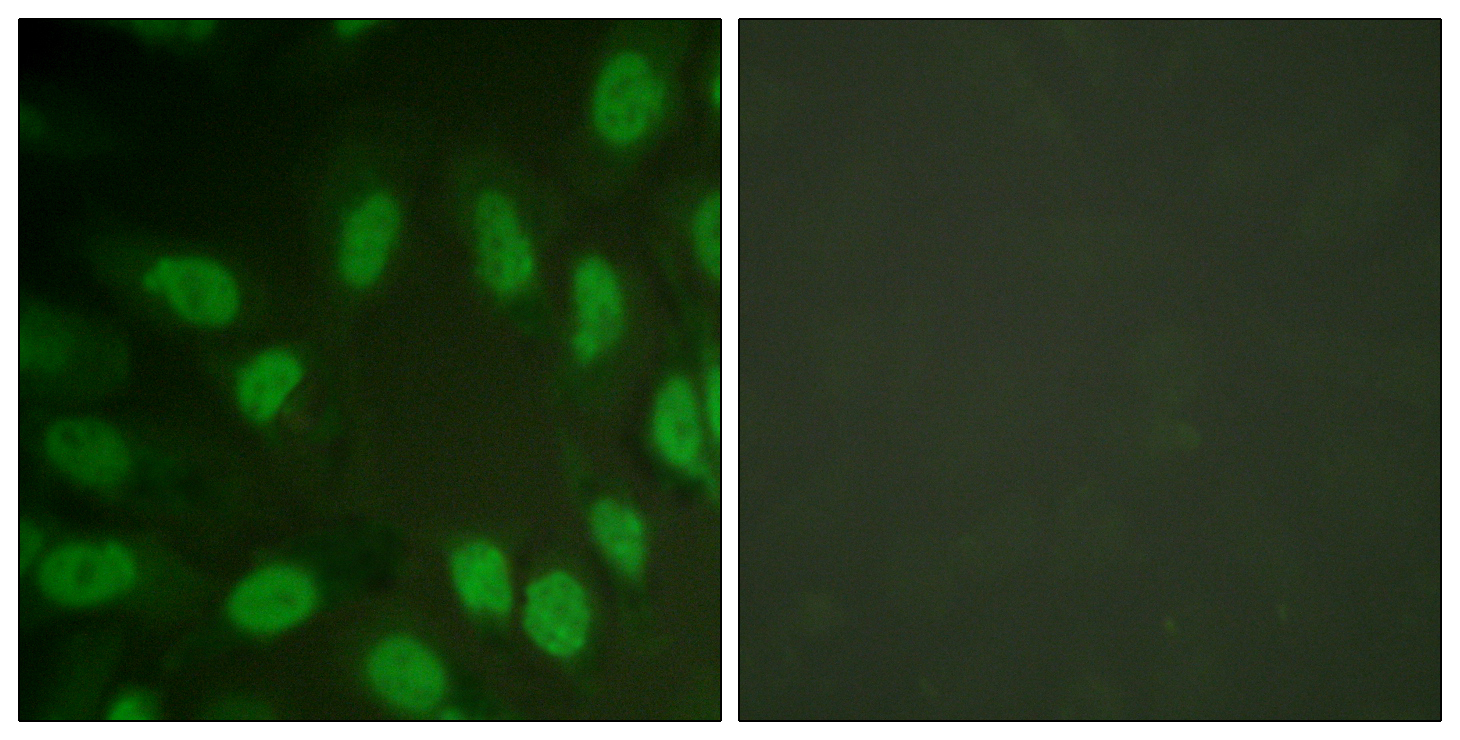
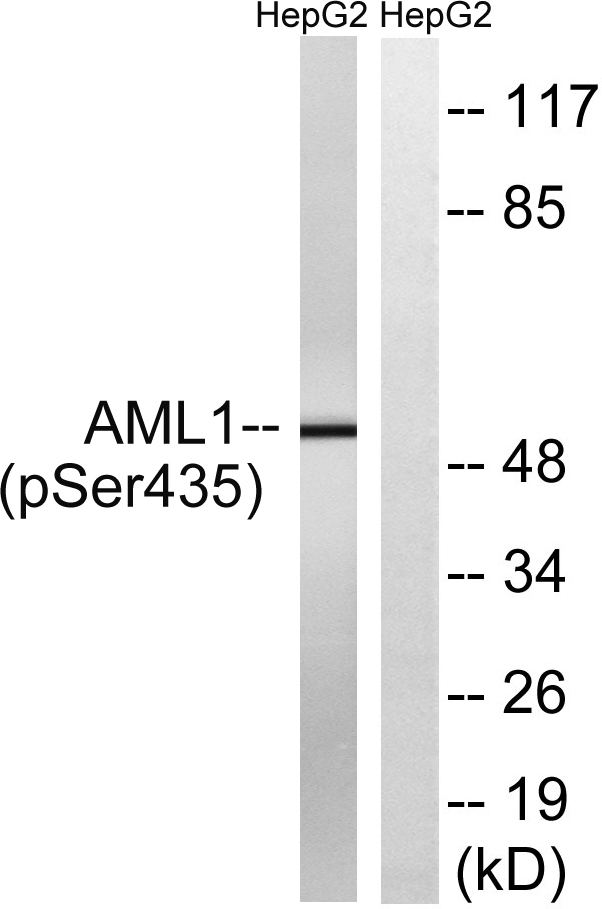
Phospho AML1 (S303) Cell-Based Colorimetric ELISA Kit
- Catalog No.:KA1235C
- Applications:ELISA
- Reactivity:Human;Mouse;Rat
- Gene Name:
- RUNX1
- Human Gene Id:
- 861
- Human Swiss Prot No:
- Q01196
- Mouse Swiss Prot No:
- Q03347
- Rat Swiss Prot No:
- Q63046
- Storage Stability:
- 2-8°C/6 months
- Other Name:
- Runt-related transcription factor 1 (Acute myeloid leukemia 1 protein) (Core-binding factor subunit alpha-2) (CBF-alpha-2) (Oncogene AML-1) (Polyomavirus enhancer-binding protein 2 alpha B subunit) (PEA2-alpha B) (PEBP2-alpha B) (SL3-3 enhancer factor 1 alpha B subunit) (SL3/AKV core-binding factor alpha B subunit)
- Detection Method:
- Colorimetric
- Background:
- alternative products:Additional isoforms seem to exist,caution:The fusion of AML1 with EAP in T-MDS induces a change of reading frame in the latter resulting in 17 AA unrelated to those of EAP.,disease:A chromosomal aberration involving RUNX1/AML1 is a cause of chronic myelogenous leukemia (CML). Translocation t(3;21)(q26;q22) with EAP, MSD1 or EVI1.,disease:A chromosomal aberration involving RUNX1/AML1 is a cause of chronic myelomonocytic leukemia. Inversion inv(21)(q21;q22) with USP16.,disease:A chromosomal aberration involving RUNX1/AML1 is a cause of M2 type acute myeloid leukemia (AML-M2). Translocation t(8;21)(q22;q22) with RUNX1T1/MTG8/ETO.,disease:A chromosomal aberration involving RUNX1/AML1 is a cause of therapy-related myelodysplastic syndrome (T-MDS). Translocation t(3;21)(q26;q22) with EAP, MSD1 or EVI1.,disease:A chromosomal aberration involving RUNX1/AML1 is found in childhood acute lymphoblastic leukemia (ALL). Translocation t(12;21)(p13;q22) with TEL. The translocation fuses the 3'-end of TEL to the alternate 5'-exon of AML-1H.,disease:A chromosomal aberration involving RUNX1/AML1 is found in therapy-related myeloid malignancies. Translocation t(16;21)(q24;q22) that forms a RUNX1-CBFA2T3 fusion protein.,disease:Defects in RUNX1 are the cause of familial platelet disorder with associated myeloid malignancy (FPDMM) [MIM:601399]. FPDMM is an autosomal dominant disease characterized by qualitative and quantitative platelet defects, and propensity to develop acute myelogenous leukemia.,domain:A proline/serine/threonine rich region at the C-terminus is necessary for transcriptional activation of target genes.,function:CBF binds to the core site, 5'-PYGPYGGT-3', of a number of enhancers and promoters, including murine leukemia virus, polyomavirus enhancer, T-cell receptor enhancers, LCK, IL-3 and GM-CSF promoters. The alpha subunit binds DNA and appears to have a role in the development of normal hematopoiesis. Isoform AML-1L interferes with the transactivation activity of RUNX1. Acts synergistically with ELF4 to transactivate the IL-3 promoter and with ELF2 to transactivate the mouse BLK promoter. Inhibits MYST4-dependent transcriptional activation.,PTM:Methylated.,PTM:Phosphorylated in its C-terminus upon IL-6 treatment. Phosphorylation enhances interaction with MYST3.,similarity:Contains 1 Runt domain.,subunit:Heterodimer with CBFB. RUNX1 binds DNA as a monomer and through the Runt domain. DNA-binding is increased by heterodimerization. Isoform AML-1L can neither bind DNA nor heterodimerize. Interacts with TLE1 and THOC4. Interacts with ELF1, ELF2 and SPI1. Interacts via its Runt domain with the ELF4 N-terminal region. Interaction with ELF2 isoform 2 (NERF-1a) may act to repress RUNX1-mediated transactivation. Interacts with MYST3 and MYST4. Interacts with SUV39H1, leading to abrogate the transactivating and DNA-binding properties of RUNX1.,tissue specificity:Expressed in all tissues examined except brain and heart. Highest levels in thymus, bone marrow and peripheral blood.,
- Function:
- immune system development, regulation of myeloid leukocyte differentiation, negative regulation of myeloid leukocyte differentiation, positive regulation of myeloid leukocyte differentiation, transcription, regulation of transcription, DNA-dependent, regulation of transcription from RNA polymerase II promoter, positive regulation of biosynthetic process,positive regulation of macromolecule biosynthetic process, positive regulation of macromolecule metabolic process,positive regulation of gene expression, hemopoiesis, myeloid cell differentiation, regulation of granulocyte differentiation, negative regulation of granulocyte differentiation, positive regulation of granulocyte differentiation,positive regulation of cellular biosynthetic process, regulation of transcription, negative regulation of cell differentiation, positive regulation of cell differentiation, regulation of myeloid cell d
- Subcellular Location:
- Nucleus.
- Expression:
- Expressed in all tissues examined except brain and heart. Highest levels in thymus, bone marrow and peripheral blood.
- June 19-2018
- WESTERN IMMUNOBLOTTING PROTOCOL
- June 19-2018
- IMMUNOHISTOCHEMISTRY-PARAFFIN PROTOCOL
- June 19-2018
- IMMUNOFLUORESCENCE PROTOCOL
- September 08-2020
- FLOW-CYTOMEYRT-PROTOCOL
- May 20-2022
- Cell-Based ELISA│解您多样本WB检测之困扰
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-ACETYL-PROTEIN
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-PHOSPHO-PROTEIN
- July 13-2018
- Antibody-FAQs