
-
Weak or no signal
 1Possible Cause
1Possible CauseAntibody Sensitivity And Reactivity
SuggestionPlease refer to the "Sensitivity/Specificity" section of the antibody webpage. Sensitivity to transfection or recombinant protein antibodies alone is insufficient to detect endogenous levels of the target. Endogenous sensitive antibodies are applicable to all sample types (endogenous, transfected, or recombinant proteins). Additionally, it may be necessary to increase or decrease the amount of antibodies used to obtain the optimal signal, depending on the abundance of target proteins in the sample. We suggest using the dilution ratio displayed on the product website and manual as the starting point for optimization.
Another factor to consider is the species reactivity of antibodies. CST has conducted internal validation on the species listed in the "Reactivity" section of the product webpage. If your target species are not listed, please contact our technical support team, who will be happy to help you determine the expected reactivity of the antibody with the protein sequence of your model species.
 2Possible Cause
2Possible CauseLow levels of phosphorylated or modified proteins
SuggestionIn untreated cell lines or tissues, the basal expression levels of many post-translational modified proteins are low. We recommend using PhosphoSitePlus to search for literature on the use of low throughput methods related to your specific modification site, or using our target positive control processing table to search for processing methods and cell lines or tissues that can serve as positive controls.
Adding protease and phosphatase inhibitors to cell extracts is crucial for avoiding protein degradation and maintaining protein yield. Sodium pyrophosphate (final concentration of 2.5 mM) and β - glycerophosphate (final concentration of 1.0 mM) should be added as serine/threonine phosphatase inhibitors to the lysis buffer. Sodium orthovanadate (final concentration of 2.5 mM) should be added to inhibit tyrosine phosphatase. Protease Inhibitor Cocktail (100X) (# 5871) or Protease/Phosphatase Inhibitor Cocktail (100X) (# 5872) can also be used.
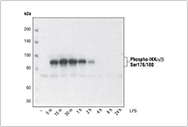
For example, inducing phosphorylation through treatment: using Phospho-IKK α/Β (Ser176/180) (16A6) Rabbit mAb for differentiation with TPA (# 9905, 80 nM, 24 hours), and performing Western blot analysis on the extract of THP-1 cells treated with 1 μ g/mL LPS for the indicated time. Phosphorylation of certain targets such as IKKa/b requires specific processing conditions to achieve optimal expression levels.
 3Possible Cause
3Possible CauseLow Protein Expression Levels in tissues or cell lines
SuggestionWe recommend using expression profiling analysis tools such as BioGPS or Human Protein Atlas, as well as scientific literature, to determine whether cells or animal tissues are expected to fully express the target protein of interest. We always recommend setting up known positive controls to assess experimental results. To learn about the recommended control information for many of our antibodies, please refer to the positive control processing table for our targets.
For whole cell extracts, it is recommended to sample at least 20-30 ug of protein per lane to detect the total amount of target/unmodified target in the whole tissue extract. However, it is usually necessary to increase the total protein loading to at least 100ug per lane to detect modified targets (such as phosphorylation and cleavage targets) in whole tissue extracts. When only a small fraction of cells in the organization contain post-translational modification targets, the whole tissue extract may need to be loaded with more proteins. Adding protease and phosphatase inhibitors to cell extracts is crucial for avoiding protein degradation and maintaining protein yield. We suggest adding leuprorelin (final concentration of 1.0ug/mL) and PMSF (# 8553) as protease inhibitors to the lysis buffer. Protease Inhibitor Cocktail (100X) (# 5871) or Protease/Phosphatase Inhibitor Cocktail (100X) (# 5872) can also be used.
 4Possible Cause
4Possible CauseReuse Pre Diluted Antibodies
SuggestionIt is not recommended to reuse diluted antibodies, as the stability of the diluted antibodies is poor and the dilution buffer stored for too long is prone to microbial or fungal contamination. We recommend always using freshly diluted antibodies for optimal results.
-
Tailing effect
 1Possible Cause
1Possible CauseReuse Pre Diluted Antibodies
SuggestionIt is not recommended to reuse diluted antibodies, as the stability of the diluted antibodies is poor and the dilution buffer stored for too long is prone to microbial or fungal contamination. We recommend always using freshly diluted antibodies for optimal results.
-
Speckle or speckled imprint
 1Possible Cause
1Possible CauseLow protein expression levels in tissues or cell lines
SuggestionWe recommend using expression profiling analysis tools such as BioGPS or Human Protein Atlas, as well as scientific literature, to determine whether cells or animal tissues are expected to fully express the target protein of interest. We always recommend setting up known positive controls to assess experimental results. To learn about the recommended control information for many of our antibodies, please refer to the positive control processing table for our targets.
For whole cell extracts, it is recommended to sample at least 20-30 ug of protein per lane to detect the total amount of target/unmodified target in the whole tissue extract. However, it is usually necessary to increase the total protein loading to at least 100ug per lane to detect modified targets (such as phosphorylation and cleavage targets) in whole tissue extracts. When only a small fraction of cells in the organization contain post-translational modification targets, the whole tissue extract may need to be loaded with more proteins. Adding protease and phosphatase inhibitors to cell extracts is crucial for avoiding protein degradation and maintaining protein yield. We suggest adding leuprorelin (final concentration of 1.0ug/mL) and PMSF (# 8553) as protease inhibitors to the lysis buffer. Protease Inhibitor Cocktail (100X) (# 5871) or Protease/Phosphatase Inhibitor Cocktail (100X) (# 5872) can also be used.
-
Multiple bands or non-specific binding
 1Possible Cause
1Possible CauseLow levels of phosphorylated or modified proteins
SuggestionIn untreated cell lines or tissues, the basal expression levels of many post-translational modified proteins are low. We recommend using PhosphoSitePlus to search for literature on the use of low throughput methods related to your specific modification site, or using our target positive control processing table to search for processing methods and cell lines or tissues that can serve as positive controls.
Adding protease and phosphatase inhibitors to cell extracts is crucial for avoiding protein degradation and maintaining protein yield. Sodium pyrophosphate (final concentration of 2.5 mM) and β - glycerophosphate (final concentration of 1.0 mM) should be added as serine/threonine phosphatase inhibitors to the lysis buffer. Sodium orthovanadate (final concentration of 2.5 mM) should be added to inhibit tyrosine phosphatase. Protease Inhibitor Cocktail (100X) (# 5871) or Protease/Phosphatase Inhibitor Cocktail (100X) (# 5872) can also be used.

For example, inducing phosphorylation through treatment: using Phospho-IKK α/Β (Ser176/180) (16A6) Rabbit mAb for differentiation with TPA (# 9905, 80 nM, 24 hours), and performing Western blot analysis on the extract of THP-1 cells treated with 1 μ g/mL LPS for the indicated time. Phosphorylation of certain targets such as IKKa/b requires specific processing conditions to achieve optimal expression levels.
-
Dark or black imprint
 1Possible Cause
1Possible CauseAntibody Sensitivity and Reactivity
SuggestionPlease refer to the "Sensitivity/Specificity" section of the antibody webpage. Sensitivity to transfection or recombinant protein antibodies alone is insufficient to detect endogenous levels of the target. Endogenous sensitive antibodies are applicable to all sample types (endogenous, transfected, or recombinant proteins). Additionally, it may be necessary to increase or decrease the amount of antibodies used to obtain the optimal signal, depending on the abundance of target proteins in the sample. We suggest using the dilution ratio displayed on the product website and manual as the starting point for optimization.
Another factor to consider is the species reactivity of antibodies. CST has conducted internal validation on the species listed in the "Reactivity" section of the product webpage. If your target species are not listed, please contact our technical support team, who will be happy to help you determine the expected reactivity of the antibody with the protein sequence of your model species.










