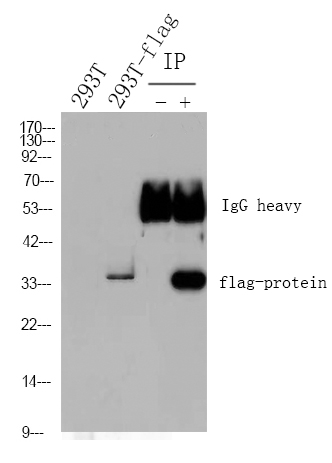
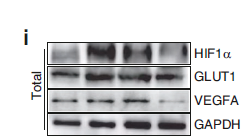
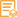Catalog: YP0667
Size
Price
Status
Qty.
200μL
$600.00
In stock
0
100μL
$340.00
In stock
0
50μL
$190.00
In stock
0
Add to cart


Collected


Collect
Main Information
Target
PAK1/2/3
Host Species
Rabbit
Reactivity
Human, Mouse, Rat
Applications
WB, IHC, IF, ELISA
MW
65kD (Observed)
Conjugate/Modification
Phospho
Detailed Information
Recommended Dilution Ratio
WB 1:500-1:2000; IHC 1:100-1:300; ELISA 1:10000; IF 1:50-200
Formulation
Liquid in PBS containing 50% glycerol, 0.5% BSA and 0.02% sodium azide.
Specificity
This antibody detects endogenous levels of PAK1/2/3 only when phosphorylated at Human:S144/141/139, Mouse:S144/141/139, Rat:S144/141/139..The name of modified sites may be influenced by many factors, such as species (the modified site was not originally found in human samples) and the change of protein sequence (the previous protein sequence is incomplete, and the protein sequence may be prolonged with the development of protein sequencing technology). When naming, we will use the "numbers" in historical reference to keep the sites consistent with the reports. The antibody binds to the following modification sequence (lowercase letters are modification sites):YMsFT
Purification
The antibody was affinity-purified from rabbit antiserum by affinity-chromatography using epitope-specific immunogen.
Storage
-15°C to -25°C/1 year(Do not lower than -25°C)
Concentration
1 mg/ml
MW(Observed)
65kD
Modification
Phospho
Clonality
Polyclonal
Isotype
IgG
Related Products
Antigen&Target Information
Immunogen:
The antiserum was produced against synthesized peptide derived from human PAK1/2/3 around the phosphorylation site of Ser144/141/139. AA range:111-160
show all
Specificity:
This antibody detects endogenous levels of PAK1/2/3 only when phosphorylated at Human:S144/141/139, Mouse:S144/141/139, Rat:S144/141/139..The name of modified sites may be influenced by many factors, such as species (the modified site was not originally found in human samples) and the change of protein sequence (the previous protein sequence is incomplete, and the protein sequence may be prolonged with the development of protein sequencing technology). When naming, we will use the "numbers" in historical reference to keep the sites consistent with the reports. The antibody binds to the following modification sequence (lowercase letters are modification sites):YMsFT
show all
Gene Name:
PAK1/PAK2/PAK3
show all
Protein Name:
Serine/threonine-protein kinase PAK 1/Serine/threonine-protein kinase PAK 2/Serine/threonine-protein kinase PAK 3
show all
Other Name:
PAK1 ;
Serine/threonine-protein kinase PAK 1 ;
Alpha-PAK ;
p21-activated kinase 1 ;
PAK-1 ;
p65-PAK ;
PAK2 ;
Serine/threonine-protein kinase PAK 2 ;
Gamma-PAK ;
PAK65 ;
S6/H4 kinase ;
p21-activated kinase 2 ;
PAK-2 ;
p58 ;
PAK3 ;
OPHN3 ;
Serine/threonine-p
Serine/threonine-protein kinase PAK 1 ;
Alpha-PAK ;
p21-activated kinase 1 ;
PAK-1 ;
p65-PAK ;
PAK2 ;
Serine/threonine-protein kinase PAK 2 ;
Gamma-PAK ;
PAK65 ;
S6/H4 kinase ;
p21-activated kinase 2 ;
PAK-2 ;
p58 ;
PAK3 ;
OPHN3 ;
Serine/threonine-p
show all
Database Link:
Background:
This gene encodes a family member of serine/threonine p21-activating kinases, known as PAK proteins. These proteins are critical effectors that link RhoGTPases to cytoskeleton reorganization and nuclear signaling, and they serve as targets for the small GTP binding proteins Cdc42 and Rac. This specific family member regulates cell motility and morphology. Alternatively spliced transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Apr 2010],
show all
Function:
Catalytic activity:ATP + a protein = ADP + a phosphoprotein.,cofactor:Magnesium.,enzyme regulation:Activated by binding small G proteins. Binding of GTP-bound CDC42 or RAC1 to the autoregulatory region releases monomers from the autoinhibited dimer, enables phosphorylation of Thr-423 and allows the kinase domain to adopt an active structure. Also activated by binding to GTP-bound CDC42, independent of the phosphorylation state of Thr-423. Phosphorylation of Thr-84 by OXSR1 inhibits this activation.,Function:The activated kinase acts on a variety of targets. Likely to be the GTPase effector that links the Rho-related GTPases to the JNK MAP kinase pathway. Activated by CDC42 and RAC1. Involved in dissolution of stress fibers and reorganization of focal complexes. Involved in regulation of microtubule biogenesis through phosphorylation of TBCB. Activity is inhibited in cells undergoing apoptosis, potentially due to binding of CDC2L1 and CDC2L2.,PTM:Autophosphorylated when activated by CDC42/p21 and RAC1.,similarity:Belongs to the protein kinase superfamily. STE Ser/Thr protein kinase family. STE20 subfamily.,similarity:Contains 1 CRIB domain.,similarity:Contains 1 protein kinase domain.,subcellular location:Recruited to focal adhesions upon activation.,subunit:Homodimer in its autoinhibited state. Active as monomer. Interacts tightly with GTP-bound but not GDP-bound CDC42/P21 and RAC1. Binds to the caspase-cleaved p110 isoform of CDC2L1 and CDC2L2, p110C, but not the full-length proteins. Component of cytoplasmic complexes, which also contain PXN, ARHGEF6 and GIT1. Interacts with ARHGEF7. Also interacts with CRIPAK. Interacts with NISCH.,
show all
Cellular Localization:
Cytoplasm . Cell junction, focal adhesion . Cell projection, lamellipodium . Cell membrane . Cell projection, ruffle membrane . Cell projection, invadopodium . Nucleus, nucleoplasm . Chromosome . Cytoplasm, cytoskeleton, microtubule organizing center, centrosome . Colocalizes with RUFY3, F-actin and other core migration components in invadopodia at the cell periphery (PubMed:25766321). Recruited to the cell membrane by interaction with CDC42 and RAC1. Recruited to focal adhesions upon activation. Colocalized with CIB1 within membrane ruffles during cell spreading upon readhesion to fibronectin. Upon DNA damage, translocates to the nucleoplasm when phosphorylated at Thr-212 where is co-recruited with MORC2 on damaged chromatin (PubMed:23260667). Localization to the centrosome does not depend upon the presence of gamma-tubulin (PubMed:27012601). Localization of the active, but not inactive, protein to the adhesions and edge of lamellipodia is mediated by interaction with GIT1 (PubMed:11896197). .
show all
Tissue Expression:
Overexpressed in gastric cancer cells and tissues (at protein level) (PubMed:25766321).
show all
Research Areas:
>>MAPK signaling pathway ;
>>ErbB signaling pathway ;
>>Ras signaling pathway ;
>>cAMP signaling pathway ;
>>Chemokine signaling pathway ;
>>Axon guidance ;
>>Hippo signaling pathway - multiple species ;
>>Focal adhesion ;
>>C-type lectin receptor signaling pathway ;
>>Natural killer cell mediated cytotoxicity ;
>>T cell receptor signaling pathway ;
>>Fc gamma R-mediated phagocytosis ;
>>Regulation of actin cytoskeleton ;
>>Epithelial cell signaling in Helicobacter pylori infection ;
>>Pathogenic Escherichia coli infection ;
>>Salmonella infection ;
>>Human immunodeficiency virus 1 infection ;
>>Proteoglycans in cancer ;
>>Renal cell carcinoma
>>ErbB signaling pathway ;
>>Ras signaling pathway ;
>>cAMP signaling pathway ;
>>Chemokine signaling pathway ;
>>Axon guidance ;
>>Hippo signaling pathway - multiple species ;
>>Focal adhesion ;
>>C-type lectin receptor signaling pathway ;
>>Natural killer cell mediated cytotoxicity ;
>>T cell receptor signaling pathway ;
>>Fc gamma R-mediated phagocytosis ;
>>Regulation of actin cytoskeleton ;
>>Epithelial cell signaling in Helicobacter pylori infection ;
>>Pathogenic Escherichia coli infection ;
>>Salmonella infection ;
>>Human immunodeficiency virus 1 infection ;
>>Proteoglycans in cancer ;
>>Renal cell carcinoma
show all
Signaling Pathway
Cellular Processes >> Cellular community - eukaryotes >> Focal adhesion
Cellular Processes >> Cell motility >> Regulation of actin cytoskeleton
Organismal Systems >> Immune system >> Natural killer cell mediated cytotoxicity
Organismal Systems >> Immune system >> T cell receptor signaling pathway
Organismal Systems >> Immune system >> Fc gamma R-mediated phagocytosis
Organismal Systems >> Immune system >> Chemokine signaling pathway
Human Diseases >> Cancer: specific types >> Renal cell carcinoma
Environmental Information Processing >> Signal transduction >> MAPK signaling pathway
Environmental Information Processing >> Signal transduction >> ErbB signaling pathway
Environmental Information Processing >> Signal transduction >> Ras signaling pathway
Environmental Information Processing >> Signal transduction >> Hippo signaling pathway - multiple species
Environmental Information Processing >> Signal transduction >> cAMP signaling pathway
Reference Citation({{totalcount}})
Catalog: YP0667
Size
Price
Status
Qty.
200μL
$600.00
In stock
0
100μL
$340.00
In stock
0
50μL
$190.00
In stock
0
Add to cart


Collected


Collect
Recently Viewed Products
Clear allPRODUCTS
CUSTOMIZED
ABOUT US
Toggle night Mode
{{pinfoXq.title || ''}}
Catalog: {{pinfoXq.catalog || ''}}
Filter:
All
{{item.name}}
{{pinfo.title}}
-{{pinfo.catalog}}
Main Information
Target
{{pinfo.target}}
Reactivity
{{pinfo.react}}
Applications
{{pinfo.applicat}}
Conjugate/Modification
{{pinfo.coupling}}/{{pinfo.modific}}
MW (kDa)
{{pinfo.mwcalc}}
Host Species
{{pinfo.hostspec}}
Isotype
{{pinfo.isotype}}
Product {{index}}/{{pcount}}
Prev
Next
{{pvTitle}}
Scroll wheel zooms the picture
{{pvDescr}}