Catalog: YC0073
Size
Price
Status
Qty.
200μL
$450.00
In stock
0
100μL
$280.00
In stock
0
40μL
$150.00
In stock
0
Add to cart


Collected


Collect
Main Information
Target
PARP
Host Species
Rabbit
Reactivity
Human, Mouse, Rat
Applications
WB, ELISA
MW
89kD (Observed)
Conjugate/Modification
Unmodified
Detailed Information
Recommended Dilution Ratio
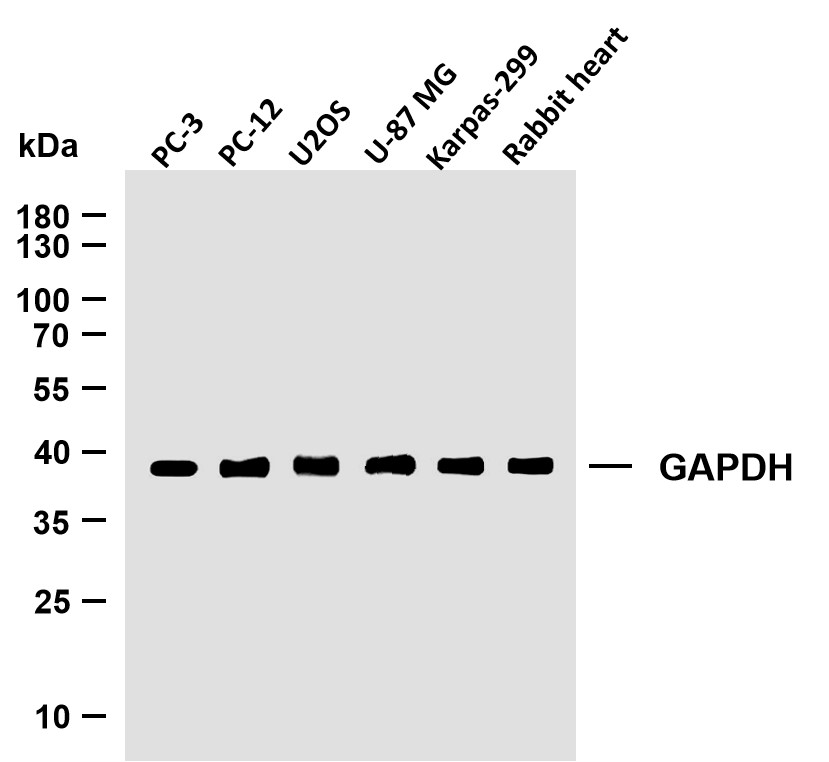
WB 1:500-1:2000; ELISA 1:5000; Not yet tested in other applications.
Formulation
Liquid in PBS containing 50% glycerol, 0.5% BSA and 0.02% sodium azide.
Specificity
Cleaved-PARP-1 (G215) Polyclonal Antibody detects endogenous levels of fragment of activated PARP-1 protein resulting from cleavage adjacent to G215.
Purification
The antibody was affinity-purified from rabbit antiserum by affinity-chromatography using epitope-specific immunogen.
Storage
-15°C to -25°C/1 year(Do not lower than -25°C)
Concentration
1 mg/ml
MW(Observed)
89kD
Modification
Unmodified
Clonality
Polyclonal
Isotype
IgG
Related Products
Antigen&Target Information
Immunogen:
The antiserum was produced against synthesized peptide derived from human PARP. AA range:196-245
show all
Specificity:
Cleaved-PARP-1 (G215) Polyclonal Antibody detects endogenous levels of fragment of activated PARP-1 protein resulting from cleavage adjacent to G215.
show all
Gene Name:
PARP1
show all
Protein Name:
Poly [ADP-ribose] polymerase 1
show all
Other Name:
PARP1 ;
ADPRT ;
PPOL ;
Poly [ADP-ribose] polymerase 1 ;
PARP-1 ;
ADP-ribosyltransferase diphtheria toxin-like 1 ;
ARTD1 ;
NAD ;
+ ;
ADP-ribosyltransferase 1 ;
ADPRT 1 ;
Poly[ADP-ribose] synthase 1
ADPRT ;
PPOL ;
Poly [ADP-ribose] polymerase 1 ;
PARP-1 ;
ADP-ribosyltransferase diphtheria toxin-like 1 ;
ARTD1 ;
NAD ;
+ ;
ADP-ribosyltransferase 1 ;
ADPRT 1 ;
Poly[ADP-ribose] synthase 1
show all
Background:
This gene encodes a chromatin-associated enzyme, poly(ADP-ribosyl)transferase, which modifies various nuclear proteins by poly(ADP-ribosyl)ation. The modification is dependent on DNA and is involved in the regulation of various important cellular processes such as differentiation, proliferation, and tumor transformation and also in the regulation of the molecular events involved in the recovery of cell from DNA damage. In addition, this enzyme may be the site of mutation in Fanconi anemia, and may participate in the pathophysiology of type I diabetes. [provided by RefSeq, Jul 2008],
show all
Function:
Catalytic activity:NAD(+) + (ADP-D-ribosyl)(n)-acceptor = nicotinamide + (ADP-D-ribosyl)(n+1)-acceptor.,Function:Involved in the base excision repair (BER) pathway, by catalyzing the poly(ADP-ribosyl)ation of a limited number of acceptor proteins involved in chromatin architecture and in DNA metabolism. This modification follows DNA damages and appears as an obligatory step in a detection/signaling pathway leading to the reparation of DNA strand breaks.,miscellaneous:The ADP-D-ribosyl group of NAD(+) is transferred to an acceptor carboxyl group on a histone or the enzyme itself, and further ADP-ribosyl groups are transferred to the 2'-position of the terminal adenosine moiety, building up a polymer with an average chain length of 20-30 units.,PTM:Phosphorylated by PRKDC. Phosphorylated upon DNA damage, probably by ATM or ATR.,PTM:Poly-ADP-ribosylated by PARP2.,similarity:Contains 1 BRCT domain.,similarity:Contains 1 PARP alpha-helical domain.,similarity:Contains 1 PARP catalytic domain.,similarity:Contains 2 PARP-type zinc fingers.,subunit:Component of a base excision repair (BER) complex, containing at least XRCC1, PARP2, POLB and LIG3. Homo- and heterodimer with PARP2. Interacts with PARP3, APTX and SRY. The SWAP complex consists of NPM1, NCL, PARP1 and SWAP70. Interacts with TIAM2 and ZNF423.,
show all
Cellular Localization:
Nucleus . Nucleus, nucleolus . Chromosome . Localizes to sites of DNA damage. .
show all
Tissue Expression:
Research Areas:
>>Base excision repair ;
>>NF-kappa B signaling pathway ;
>>Apoptosis ;
>>Necroptosis ;
>>Diabetic cardiomyopathy
>>NF-kappa B signaling pathway ;
>>Apoptosis ;
>>Necroptosis ;
>>Diabetic cardiomyopathy
show all
Signaling Pathway
Reference Citation({{totalcount}})
Catalog: YC0073
Size
Price
Status
Qty.
200μL
$450.00
In stock
0
100μL
$280.00
In stock
0
40μL
$150.00
In stock
0
Add to cart


Collected


Collect
Recently Viewed Products
Clear allPRODUCTS
CUSTOMIZED
ABOUT US
Toggle night Mode
{{pinfoXq.title || ''}}
Catalog: {{pinfoXq.catalog || ''}}
Filter:
All
{{item.name}}
{{pinfo.title}}
-{{pinfo.catalog}}
Main Information
Target
{{pinfo.target}}
Reactivity
{{pinfo.react}}
Applications
{{pinfo.applicat}}
Conjugate/Modification
{{pinfo.coupling}}/{{pinfo.modific}}
MW (kDa)
{{pinfo.mwcalc}}
Host Species
{{pinfo.hostspec}}
Isotype
{{pinfo.isotype}}
Product {{index}}/{{pcount}}
Prev
Next
{{pvTitle}}
Scroll wheel zooms the picture
{{pvDescr}}