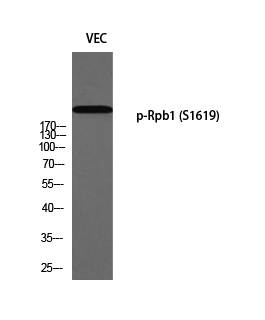
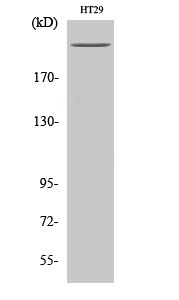Total POLR2A Cell-Based Colorimetric ELISA Kit
- Catalog No.:KA4249C
- Applications:ELISA
- Reactivity:Human;Mouse;Rat
- Gene Name:
- POLR2A
- Human Gene Id:
- 5430
- Human Swiss Prot No:
- P24928
- Mouse Swiss Prot No:
- P08775
- Storage Stability:
- 2-8°C/6 months
- Other Name:
- DNA-directed RNA polymerase II subunit RPB1 (RNA polymerase II subunit B1) (EC 2.7.7.6) (DNA-directed RNA polymerase II subunit A) (DNA-directed RNA polymerase III largest subunit) (RNA-directed RNA polymerase II subunit RPB1) (EC 2.7.7.48)
- Detection Method:
- Colorimetric
- Background:
- catalytic activity:Nucleoside triphosphate + RNA(n) = diphosphate + RNA(n+1).,function:DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates. Largest and catalytic component of RNA polymerase II which synthesizes mRNA precursors and many functional non-coding RNAs. Forms the polymerase active center together with the second largest subunit. Pol II is the central component of the basal RNA polymerase II transcription machinery. It is composed of mobile elements that move relative to each other. RPB1 is part of the core element with the central large cleft, the clamp element that moves to open and close the cleft and the jaws that are thought to grab the incoming DNA template. At the start of transcription, a single stranded DNA template strand of the promoter is positioned within the central active site cleft of Pol II. A bridging helix emanates from RPB1 and crosses the cleft near the catalytic site and is thought to promote translocation of Pol II by acting as a ratchet that moves the RNA-DNA hybrid through the active site by switching from straight to bent conformations at each step of nucleotide addition. During transcription elongation, Pol II moves on the template as the transcript elongates. Elongation is influenced by the phosphorylation status of the C-terminal domain (CTD) of Pol II largest subunit (RPB1), which serves as a platform for assembly of factors that regulate transcription initiation, elongation, termination and mRNA processing. Acts as a RNA-dependent RNA polymerase when associated with small delta antigen of Hepatitis delta virus, acting both as a replicate and transcriptase for the viral RNA circular genome.,miscellaneous:The binding of ribonucleoside triphosphate to the RNA polymerase II transcribing complex probably involves a two-step mechanism. The initial binding seems to occur at the entry (E) site and involves a magnesium ion temporarily coordinated by three conserved aspartate residues of the two largest RNA Pol II subunits. The ribonucleoside triphosphate is transferred by a rotation to the nucelotide addition (A) site for pairing with the template DNA. The catalytic A site involves three conserved aspartate residues of the RNA Pol II largest subunit which permanently coordinate a second magnesium ion.,PTM:The tandem 7 residues repeats in the C-terminal domain (CTD) can be highly phosphorylated. The phosphorylation activates Pol II. Phosphorylation occurs mainly at residues 'Ser-2' and 'Ser-5' of the heptapepdtide repeat. The phosphorylation state is believed to result from the balanced action of site-specific CTD kinases and phosphataes, and a "CTD code" that specifies the position of Pol II within the transcription cycle has been proposed.,similarity:Belongs to the RNA polymerase beta' chain family.,similarity:Contains 1 C2H2-type zinc finger.,subunit:Component of the RNA polymerase II (Pol II) complex consisting of 12 subunits (By similarity). The phosphorylated C-terminal domain interacts with FNBP3 and SYNCRIP. Interacts with SAFB/SAFB1. Interacts with CCNL1 and MYO1C (By similarity). Interacts with CCNL2 and SFRS19. Component of a complex which is at least composed of HTATSF1/Tat-SF1, the P-TEFb complex components CDK9 and CCNT1, RNA polymerase II, SUPT5H, and NCL/nucleolin. Interacts with PAF1.,
- Function:
- RNA splicing, via transesterification reactions, RNA splicing, via transesterification reactions with bulged adenosine as nucleophile, nuclear mRNA splicing, via spliceosome, transcription, transcription, DNA-dependent, transcription initiation, RNA elongation, regulation of transcription, DNA-dependent, transcription from RNA polymerase II promoter,transcription initiation from RNA polymerase II promoter, RNA elongation from RNA polymerase II promoter, RNA processing, mRNA processing, protein complex assembly, RNA splicing, mRNA metabolic process, RNA biosynthetic process, macromolecular complex subunit organization, regulation of transcription, regulation of RNA metabolic process, macromolecular complex assembly, protein complex biogenesis,
- Subcellular Location:
- Nucleus . Cytoplasm . Chromosome . Hypophosphorylated form is mainly found in the cytoplasm, while the hyperphosphorylated and active form is nuclear (PubMed:26566685). Co-localizes with kinase SRPK2 and helicase DDX23 at chromatin loci where unscheduled R-loops form (PubMed:28076779). .
- June 19-2018
- WESTERN IMMUNOBLOTTING PROTOCOL
- June 19-2018
- IMMUNOHISTOCHEMISTRY-PARAFFIN PROTOCOL
- June 19-2018
- IMMUNOFLUORESCENCE PROTOCOL
- September 08-2020
- FLOW-CYTOMEYRT-PROTOCOL
- May 20-2022
- Cell-Based ELISA│解您多样本WB检测之困扰
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-ACETYL-PROTEIN
- July 13-2018
- CELL-BASED-ELISA-PROTOCOL-FOR-PHOSPHO-PROTEIN
- July 13-2018
- Antibody-FAQs